HIF1A-mediated pathways promote euploid cell survival in chromosomally mosaic embryos
Curation statements for this article:-
Curated by eLife
eLife Assessment
Sanchez-Vasquez et al establish an innovative approach to induce aneuploidy in preimplantation embryos. This important study extends the author's previous publications evaluating the consequences of aneuploidy in the mammalian embryo. In this work, the authors investigate the developmental potential of aneuploid embryos and characterize changes in gene expression profiles under normoxic and hypoxic culture conditions. Using a solid methodology they identify sensitivity to Hif1alpha loss in aneuploid embryos, and in further convincing experiments they assess how levels of DNA damage and DNA repair are altered under hypoxic and normoxic conditions.
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
- Evaluated articles (preLights)
Abstract
Human fertility is suboptimal in part by error-prone divisions during early cleavage stages, which frequently result in chromosomal aneuploidy. Most human pre-implantation embryos are mosaics of euploid and aneuploid cells, yet those with a low proportion of aneuploid cells can develop to term at rates similar to fully euploid embryos. How embryos manage aneuploidy during early development remains poorly understood – yet this knowledge is crucial for improving fertility outcomes and reducing developmental defects. To investigate these mechanisms, we established a new mouse model of chromosome mosaicism to trace the fate of aneuploid cells during pre-implantation development. We previously used the Mps1 inhibitor reversine to induce aneuploidy. Here, we demonstrate that the more specific Mps1 inhibitor AZ3146 similarly disrupts chromosome segregation but supports higher developmental potential than reversine. AZ3146-treated embryos transiently upregulate hypoxia-inducible factor-1A (HIF1A) without triggering Trp53 activation. Given that pre-implantation embryos develop in a hypoxic environment in vivo, we further explored the role of oxygen tension. Hypoxia exposure in vitro reduced DNA damage in response to Mps1 inhibition and increased the proportion of euploid cells in mosaic epiblast. Conversely, HIF1A inhibition decreased the proportion of aneuploid cells. Together, these findings uncover a role for hypoxia signaling in modulating the response to chromosomal errors and suggest new strategies to improve the developmental potential of mosaic human embryos.
Article activity feed
-

-

-

-

eLife Assessment
Sanchez-Vasquez et al establish an innovative approach to induce aneuploidy in preimplantation embryos. This important study extends the author's previous publications evaluating the consequences of aneuploidy in the mammalian embryo. In this work, the authors investigate the developmental potential of aneuploid embryos and characterize changes in gene expression profiles under normoxic and hypoxic culture conditions. Using a solid methodology they identify sensitivity to Hif1alpha loss in aneuploid embryos, and in further convincing experiments they assess how levels of DNA damage and DNA repair are altered under hypoxic and normoxic conditions.
-

Reviewer #1 (Public review):
Summary:
This paper developed a model of chromosome mosaicism by using a new aneuploidy-inducing drug (AZ3146), and compared this to their previous work where they used reversine, to demonstrate the fate of aneuploid cells during murine preimplantation embryo development. They found that AZ3146 acts similarly to reversine in inducing aneuploidy in embryos, but interestingly showed that the developmental potential of embryos is higher in AZ3146-treated vs. reversine-treated embryos. This difference was associated with changes in HIF1A, p53 gene regulation, DNA damage, and fate of euploid and aneuploid cells when embryos were cultured in a hypoxic environment.
Strengths:
In the current study, the authors investigate the fate of aneuploid cells in the preimplantation murine embryo using a specific …
Reviewer #1 (Public review):
Summary:
This paper developed a model of chromosome mosaicism by using a new aneuploidy-inducing drug (AZ3146), and compared this to their previous work where they used reversine, to demonstrate the fate of aneuploid cells during murine preimplantation embryo development. They found that AZ3146 acts similarly to reversine in inducing aneuploidy in embryos, but interestingly showed that the developmental potential of embryos is higher in AZ3146-treated vs. reversine-treated embryos. This difference was associated with changes in HIF1A, p53 gene regulation, DNA damage, and fate of euploid and aneuploid cells when embryos were cultured in a hypoxic environment.
Strengths:
In the current study, the authors investigate the fate of aneuploid cells in the preimplantation murine embryo using a specific aneuploidy-inducing compound to generate embryos that were chimeras of euploid and aneuploid cells. The strength of the work is that they investigate the developmental potential and changes in gene expression profiles under normoxic and hypoxic culture conditions. Further, they also assessed how levels of DNA damage and DNA repair are altered in these culture conditions. They also assessed the allocation of aneuploid cells to the divergent cell lineages of the blastocyst stage embryo.
-

Author response:
The following is the authors’ response to the previous reviews
Reviewer #1(Public review):
We deeply appreciate the reviewer comments on our manuscript. Following up the revisions, our manuscript has been improved thanks to their insightful remarks. We have proceeded with all the required changes.
Weaknesses:
The authors have still not addressed the inconsistent/missing description for sample size, the appropriate number of * for each figure panel, and the statistical tests used.
Description of sample size, specific P value and statistical test used has been added it both in the main text, figures and figure legends.
The authors assign 5% oxygen as hypoxia. This is not the case as the in vivo environment is close to this value. 5% is normoxia. Clinical IVF/embryo culture occurs at 5% O2. Please adjust your narrative …
Author response:
The following is the authors’ response to the previous reviews
Reviewer #1(Public review):
We deeply appreciate the reviewer comments on our manuscript. Following up the revisions, our manuscript has been improved thanks to their insightful remarks. We have proceeded with all the required changes.
Weaknesses:
The authors have still not addressed the inconsistent/missing description for sample size, the appropriate number of * for each figure panel, and the statistical tests used.
Description of sample size, specific P value and statistical test used has been added it both in the main text, figures and figure legends.
The authors assign 5% oxygen as hypoxia. This is not the case as the in vivo environment is close to this value. 5% is normoxia. Clinical IVF/embryo culture occurs at 5% O2. Please adjust your narrative around this.
We define in our manuscript “normoxia” as the standard atmospheric oxygen levels in tissue culture incubators, which range from about 20–21% oxygen. Our definition of hypoxia is 5% concentration of oxygen, taking into consideration the standard levels of oxygen in the IVF clinics. Physiological oxygen in mouse varies from ~1.5% to 8% (Alva et al 2022). Considering that these levels of oxygen are the standard levels in tissue culture practices, a paragraph has been added to the discussion and materials and methods for further clarification
Reviewer #2 (Public review):
Weakness:
Given that this is a study on the induction of aneuploidy, it would be meaningful to assess aneuploidy immediately after induction, and then again before implantation. This is also applicable to the competition experiments on page 7/8. What is shown is the competitiveness of treated cells. Because the publication centers around aneuploidy, inclusion of such data in the main figure at all relevant points would strengthen it. There is some evaluation of karyotypes only in the supplemental - why? Would be good not to rely on a single assay that the authors appear to not give much importance.
This is an excellent point. However, due to the stochasticity of the arising of aneuploidies when embryos are treated with AZ3146 and reversine (Bolton et al 2016), every treatment is likely to generate different levels of aneuploidy. Due to this, and to the technical limitations of generating single-cell genomic DNA sequencing at the blastocyst stage, we were unable to determine the karyotype of all cells after different conditions. Nevertheless, Regin et al 2024 (eLife) showed similar results on the overall transcriptome changes of different dosages of aneuploidy: high dosage embryos overexpress p53, like reversine-treated embryos; meanwhile, low dosage embryos overexpress the hypoxic pathway, including HIF1A, similar to embryos treated with AZ3146.
Reviewer #1 (Recommendations for the authors):
Corrections required before final publishing:
Please ensure that the number of asterisks is in alignment with standard convention (* <0.05; ** <0.01; *** <0.001; **** <0.0001). If you want to describe an exact P -vale it should be presented as P = 0.0004. line 108 *** is <0.0004. line 263 * P<0.0044
Same issue appears in lines 697, 711, 722, 753, 685
Specific values have been added in the figures and modified in the text.
Line 199: "...viable E9.5 embryos" missing "Figure S1D"
Modified in manuscript
Line 120: "...decidua" please add "Figure S1C"
Modified in manuscript
Line 126-127: Please add a description for the results (morula) in Fig 1D, e.g., It appears that YH2Ax persists from 8-cell to morula when treated with Reversine but not AZ3146"
At the morula stage, the levels of γH2A.X in reversine- and AZ3146-treated embryos are similar (Fig. 1E). However, at the blastocyst stage, high levels of γH2A.X are maintained in reversine-treated embryos and reduced in AZ3146-treated embryos, suggesting some level of DNA repair between the morula-to-blastocyst stages (Fig. S2A). In contrast, in hypoxia, the levels of γH2A.X are low in the three treatments at the morula stages, suggesting that DNA repair can be enhanced under hypoxic conditions. Similar results have been reported in somatic cells (Marti et al., 2021; Pietrzak et al., 2018).
Line 213: PARP1 levels were also similar under all conditions; but Fig3E, top right shows PARP1 was significantly lower with Reversine treatment; also please correct me if i am wrong, but does the phrase "all conditions" cross reference yH2AX and PARP1 between Fig 3 and Fig 1 to show the impact of hypoxia? Because from my understanding Fig 1 was done in 20% oxygen, but Fig 3 was done in 5% oxygen – hypoxia.
This is correct. Modification in the manuscript has been performed for clarification
Line 264: extra forward dash? "Reversine/AZ3146/ aggregation"
Modified in manuscript
Line 644: you don't have a control for IDF treatment, so how did you differentiate between impact of aneuploid drugs vs IDF treatment alone? Would the impact observed be due to compounding effect of aneuploidy drugs + IDF?
This is a great observation. We previously demonstrated that IDF-1174 treatments in embryos do not affect pre-implantation development (Fig. S3).
Line 681: change their behaviour is a vague statement. Be specific.
Modified in manuscript
Line 676 missing bracket "E)"
Modified in manuscript
Line 680: "...significantly on" should be "for"
Modified in manuscript
Line 682-685: "...hypoxia favours the survival of reversine-induced aneuploid cells." does it? the statement before this says in Rev/AZ chimeras, AZ blastomeres contribute similarly to reversine-blastomeres to the TE and PE but significantly increase contributions to the EPI.Wouldn't this mean hypoxia favours survival of AZ aneuploid cells in EPI?
In normoxic conditions, AZ3146 treated cells in Rev/AZ chimeras contributed mostly to the EPI and TE but not PE. In contrast, in normoxic conditions, Rev-treated cells contributed similarly to all the lineages. This result seems to be due to a better survival of Rev-treated cells under normoxic conditions (Fig. 4D-E)
Line 720: (b) shows blastocyst staining from what group? DMSO? Rev/AZ? Or are the 3 blastocysts shown here, 3 separate examples of Reversine-treated blastocysts? Would require labelling Fig S2B, and adding a short description in the corresponding figure legend
Figure (B) shows the expression pattern of PARP1 at the blastocyst stage. Modified in manuscript
Figure 2, Figure S3 and Figure S6: were these experiments performed at 5% or 20% O2, please add detail.
Modified in manuscript
Reviewer #2 (Recommendations for the authors):
Lines 45-46 understanding of reduction of aneuploidy should mention/discuss the paper of attrition/selection, of the kind by the Brivanlou lab for instance, or others. As well as allocation to specific lineages, including the authors' work.
A section in the discussion has been added in response to this recommendation. Comparison between models is debatable.
The response does not clarify whether other papers were cited instead, or the authors own work that has shown preferential allocation to TE.
-

-

eLife Assessment
Sanchez-Vasquez et al establish an innovative approach to induce aneuploidy in preimplantation embryos. This important study extends the author's previous publications evaluating the consequences of aneuploidy in the mammalian embryo. In this work, the authors investigate the developmental potential of aneuploid embryos and characterize changes in gene expression profiles under normoxic and hypoxic culture conditions. Using a solid methodology they identify sensitivity to Hif1alpha loss in aneuploid embryos, and in further convincing experiments they assess how levels of DNA damage and DNA repair are altered under hypoxic and normoxic conditions.
-

Reviewer #1 (Public review):
Summary:
This paper developed a model of chromosome mosaicism by using a new aneuploidy-inducing drug (AZ3146), and compared this to their previous work where they used reversine, to demonstrate the fate of aneuploid cells during murine preimplantation embryo development. They found that AZ3146 acts similarly to reversine in inducing aneuploidy in embryos, but interestingly showed that the developmental potential of embryos is higher in AZ3146-treated vs. reversine-treated embryos. This difference was associated with changes in HIF1A, p53 gene regulation, DNA damage, and fate of euploid and aneuploid cells when embryos were cultured in a hypoxic environment.
Strengths:
In the current study, the authors investigate the fate of aneuploid cells in the preimplantation murine embryo using a specific …
Reviewer #1 (Public review):
Summary:
This paper developed a model of chromosome mosaicism by using a new aneuploidy-inducing drug (AZ3146), and compared this to their previous work where they used reversine, to demonstrate the fate of aneuploid cells during murine preimplantation embryo development. They found that AZ3146 acts similarly to reversine in inducing aneuploidy in embryos, but interestingly showed that the developmental potential of embryos is higher in AZ3146-treated vs. reversine-treated embryos. This difference was associated with changes in HIF1A, p53 gene regulation, DNA damage, and fate of euploid and aneuploid cells when embryos were cultured in a hypoxic environment.
Strengths:
In the current study, the authors investigate the fate of aneuploid cells in the preimplantation murine embryo using a specific aneuploidy-inducing compound to generate embryos that were chimeras of euploid and aneuploid cells. The strength of the work is that they investigate the developmental potential and changes in gene expression profiles under normoxic and hypoxic culture conditions. Further, they also assessed how levels of DNA damage and DNA repair are altered in these culture conditions. They also assessed the allocation of aneuploid cells to the divergent cell lineages of the blastocyst stage embryo.
Weaknesses:
The authors have still not addressed the inconsistent/missing description for sample size, the appropriate number of * for each figure panel, and the statistical tests used.
The authors assign 5% oxygen as hypoxia. This is not the case as the in vivo environment is close to this value. 5% is normoxia. Clinical IVF/embryo culture occurs at 5% O2. Please adjust your narrative around this.
-

Reviewer #2 (Public review):
Summary:
This study by Sanchez-Vasquez is a very innovative approach to induce aneuploidy and then study the contribution of treated cells to different lineages, including post implantation. It connects well to the authors previous work to induce mosaic aneuploidies. The authors identify sensitivity to HIF1a loss in treated embryos with likely aneuploidy. This work is part of an important line of work with evaluates the consequences of aneuploidy in mammalian embryo.
Weaknesses:
Given that this is a study on the induction of aneuploidy, it would be meaningful to assess aneuploidy immediately after induction, and then again before implantation. This is also applicable to the competition experiments on page 7/8. What is shown is the competitiveness of treated cells. Because the publication centers around …
Reviewer #2 (Public review):
Summary:
This study by Sanchez-Vasquez is a very innovative approach to induce aneuploidy and then study the contribution of treated cells to different lineages, including post implantation. It connects well to the authors previous work to induce mosaic aneuploidies. The authors identify sensitivity to HIF1a loss in treated embryos with likely aneuploidy. This work is part of an important line of work with evaluates the consequences of aneuploidy in mammalian embryo.
Weaknesses:
Given that this is a study on the induction of aneuploidy, it would be meaningful to assess aneuploidy immediately after induction, and then again before implantation. This is also applicable to the competition experiments on page 7/8. What is shown is the competitiveness of treated cells. Because the publication centers around aneuploidy, inclusion of such data in the main figure at all relevant points would strengthen it. There is some evaluation of karyotypes only in the supplemental - why? Would be good not to rely on a single assay that the authors appear to not give much importance.
-

Author response:
The following is the authors’ response to the original reviews.
We deeply appreciate the reviewer comments on our manuscript. We have proceeded with all the minor changes mentioned. We also want to emphasize three major points:
(1) Reversine has been shown to have several off-targets effects. Including inducing apoptosis (Chen et al. J Bone Oncol. 2024).
(2) Hypoxia varies from 2% to 6%. Our definition of hypoxia is 5% concentration of oxygen with 5% concentration of CO2, taking into consideration the standard levels of oxygen in the IVF clinics. Physiological oxygen in mouse varies from ~1.5% to 8%.
(3) Natale et al. 2004 (Dev Bio) and Sozen et al. 2015 (Mech of Dev) described that inhibition of p38 deeply affect the development of pre-implantation embryos after the 8-cell stage. For this reason, comprehensible dissect …
Author response:
The following is the authors’ response to the original reviews.
We deeply appreciate the reviewer comments on our manuscript. We have proceeded with all the minor changes mentioned. We also want to emphasize three major points:
(1) Reversine has been shown to have several off-targets effects. Including inducing apoptosis (Chen et al. J Bone Oncol. 2024).
(2) Hypoxia varies from 2% to 6%. Our definition of hypoxia is 5% concentration of oxygen with 5% concentration of CO2, taking into consideration the standard levels of oxygen in the IVF clinics. Physiological oxygen in mouse varies from ~1.5% to 8%.
(3) Natale et al. 2004 (Dev Bio) and Sozen et al. 2015 (Mech of Dev) described that inhibition of p38 deeply affect the development of pre-implantation embryos after the 8-cell stage. For this reason, comprehensible dissect the interaction between p53, HIF1A and p38 during aneuploid stress is challenging. We do not discard a double function of p38 during lineage specification and in response to DNA damage.
Reviewer #2 (Recommendations for the authors):
(1) Line 69: Please add the species used in your cited publications (murine).
Fixed
(2) Line 72: Consider changing "Because" to "As".
Fixed
(3) Line 88: "from the nuclei" - please refer to where the reader may find the example provided (Figure S1A).
Fixed
(4) Line 89: This should be Figure S1B as no quantification is presented in S1A. S1A only contains examples of micronuclei.
Fixed
(5) Line 91: Refer to Figure S1A.
Fixed
(6) Line 91-93: Are these numbers correct? The query arises from the numbers presented in Figure S1B. Please define how the median was calculated; is it micronuclei CREST+ plus micronuclei CREST-?
Fixed. We did not differentiate in these percentage the presence of CREST.
(7) Line 95: extra/missing bracket?
Fixed
(8) Line 88-91:
[a] Regarding the number of cells with micronuclei in this text, please clarify your sample size and how the percentages were calculated as they currently do not align (e.g., are these the total number of embryos from a single experimental replicate?).
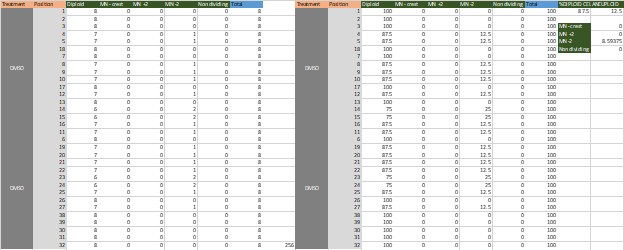
Also, different numbers are found here and in the figure legend: (DMSO-22/256 cells from 32 embryos; Rev-82/144 cells from 18 embryos; AZ-182/304 cells from 38 embryos) vs. Fig S1 legend (DMSO-n=128 cells; Rev-72 cells; AZ-152 cells).
[c] Is the median calculated using the numbers presented above? If yes, then the numbers do not tally, please check (DMSO-22/256 cells=8.6%; Rev-82/144 cells=56.9%; AZ-182/304 cells =59.9%) vs. Line 91-93: DMSO=12.5%, Rev=75%; AZ=62.5% blastomeres had micronuclei.
The percentage represents the average of aneuploidy per embryo after normalization.
See table for DMSO. This number represents the average of aneuploid cells each aneuploid embryo has. Notice that some embryos are fully diploid. Some have more that 12.5% -> 25%. Most of the aneuploid embryos have 12.5% of aneuploidy. It is not black and white as how many aneuploid cell there is in the sample but a full understanding of how aneuploid are the aneuploid embryos in each sample.
Author response image 1.
(9) Line 108:
[a] "n=28 per treatment" please clarify whether this refers to the number of embryos or cells and also add how many independent replicate experiments this data is representative of. as the text only refers to Figure 1C you can remove the P-values for ** and *.
Number of embryos. Fixed
(10) Line 111: Suggest citing Figure 1C at the end of the sentence.
Fixed
(11) Line 118-119: the reference to figures require updating to ensure they refer to the appropriate figure; ...decidua (Figure S1C)...viable E9.5 embryos (Figure S1D).
Fixed
(12) Line 126: A description of the data in Figures 1D and 1E is missing. Also, consider describing the DNA damage observed in the DMSO control group. Visually, it appears that DNA damage reduces from the 8-cell to the morula stage (Figure 1E) but increases at the blastocyst stage (Figure S2A)? Point for discussion for a normal rate of DNA damage?
Agree, there is some DNA damage in the TE in blastocyst
(13) Line 134: 8 EPI and 4 PE cells in what group?
Fixed: DMSO-treated embryos
(14) Line 137: Could this also suggest that AZ and reversine induce DNA damage through a different mechanism/pathway, resulting in the differential impact observed? Despite both being inhibitors of Mps1.
This is a possibility.
(15) Line 153: the legend for Figure 2A says the Welch t-test was performed, but the Mann-Whitney U-test was stated here. Which is correct?
Welch’s t-test
(16) Line 155: ...at the blastocyst stage. Compared to what?
DMSO-treated embryos
(17) Line 160: Data in Figure 2B requires the definition of P-values for *, **, ****. Please add one for **** and remove the one for ***.
Fixed
(18) Line 173-174: Data in Fig. 4 requires the definition of the P-values for ****. Please remove the others.
Fixed
(19) Line 180: Instead of jumping across figures, this section would benefit from stating the numbers directly to allow for an accurate comparison, e.g. 64 and 7 in Figure 2D vs. X and Y in Figure 1C.
(20) Line 187: Hif1a should be italicised.
Fixed
(21) Line 197: Based on the description here, I believe you are missing a reference to Figure 1A.
Fixed
(22) Line 203: Instead of jumping across figures, this section would benefit from stating the numbers directly to allow for accurate comparison, "particularly in the TE and PE" (67 vs 54; and 11 vs 6, respectively).
(23) Line 209-210:
[a] "...lowered the number of yH2AX foci..." is this a visual observation as quantification was performed for yH2AX intensity, not quantification of foci?
A description for PARP1 levels in morula stage embryos was presented ("...relatively low in morula), but not for yH2AX levels at this stage of development. Missing description?
Fixed
(24) Line 235: This sentence would benefit from being specific about the environmental conditions...eg "Under normoxia, DMSO/AZ3146-treated...",
(25) Line 238: The sentence should reference Figure 4F not 4G.
Fixed
(26) Line 242-243:
[a] "slightly increased... in the TE (49.06%) and PE (50%) but, strikingly, reduced... EPI (33.3%)" compared to what and in which figure?
Assuming you are comparing normoxia (4F) to hypoxia (4G), the numbers change for the TE (46.75% to 49.06%, respectively), EPI (42.88% to 33.3%, respectively), and PE (28.57% to 50%, respectively); yet these data were described as "strikingly different" for EPI (9.58 decrease) but only "slightly increased" for PE (21.42 increase). Suggest using appropriate adjectives to describe the results.
Fixed
(27) Line 256: It is stated in line 255 that treatment was performed at the zygote stage, yet this sentence says reversine treatment occurred at the 2-cell stage? Which is correct? Please amend appropriately. Refer to the comment below regarding adding a schematic to aid readers
Fixed
(28) Line 259: "n>27 per treatment" please clarify whether this refers to the number of embryos or cells and also add how many independent replicate experiments this data is representative of. Data in Figures S5A-B requires a definition of P-values for *, ****. Please remove for **, ***.
Fixed
(29) Line 261: AZ3146/reversine stated here, the figure shows Reversine/AZ3146. Please consider being consistent.
Fixed
(30) Line 263: "... normal morphology and cavitation (Figure S5D); however the image presented for Rev/DMSO and Rev/AZ3146 chimeras appear smaller with a distorted/weird shape when compared to DMSO/AZ. I believe the description does not match the images presented.
Fixed
(31) Line 267: "...similar results as 8-cell stage derived chimeras"; however, there is only a reference to Fig S5E which depicts 2-cell/zygote stage (see comment above for line 256 regarding required clarification of stage of treatment) derived chimeras. There is also a missing reference to Figure 4B, D, and/or F?
Fixed
(32) Line 271: add a reference to Figure S5E.
Fixed
(33) Line 283: "AZ3146/reversine" should be "Reversine/AZ3146" to match the figure.
Fixed
(34) Line 284: Figures 5E-F show both morphology and cavitation; the text should reflect this.
Fixed
(35) Line 281-285: I think this text requires editing to improve clarity. It is difficult for this reader to understand the authors' interpretation of the results....inhibiting HIF1A reduces morphology and cavitation. That's correct. However, this also diminished the contribution of AZ3146-treated cells to all 3 cell lineages; this is not quite accurate. AZ3146-treated cells were significantly reduced in total cell numbers because TE was significantly reduced. It is not appropriate to generalise this result to all 3 lineages, as EPI and TE appear to increase AZ's contribution following IDF treatment, albeit non-statistically significant.
Fixed
(36) Line 320: citation? ....reversine-treated embryos. Is this referring to your previous publication...Bolton 2016?
Fixed
(37) Line 344: missing space between 7.5 and IU.
Fixed
(38) Line 358: animal ethics approval number/code missing.
Fixed
(39) Line 397: missing space between "...previously" and "(Bermejo...".
Fixed
(40) Line 417: missing space between "...control" and "(Gu et...".
Fixed
(41) Line 421: missing space between "protocol" and "(Eakin...".
Fixed
(42) Line 427-429: Medium-grade mosaic chimeras were referred to as DMSO:AZ:Rev (3:3:2) here; but Figure 4 and associated legend says otherwise. Please amend appropriately. Were all medium mosaics generated in this manner? As I could only find Rev/AZ chimeras; my understanding of the Rev/AZ chimeras is 1:1 Rev:AZ instead of 3:2:3 DMSO:Rev:AZ.
Fixed
(43) Line 428: "reversine-treaded: please correct spelling.
Fixed
(44) Line 593: "n=28 per treatment" Please clarify whether this refers to the number of embryos or cells and also add how many independent replicate experiments this data is representative of.
Fixed
(45) Line 597: "through morula stage" when compared to what group?
DMSO-treated embryos
(46) Line 598: Data in Figure S5A-B requires the definition of P-values for **, ***, ****. Please remove for *. Please define the error bars. SEM/95% confidence interval?
Fixed
(47) Line 604-607: Regarding 2B, no statistical test is stated yet Mann-Whitney was stated in Line 160 of the results section. Please confirm which test was used and include it in both sections for consistency.
Fixed
(48) Line 608: "Chemical downregulation of HIF1A"... this is not described in the results/methods section or shown in the figure. Please amend all sections for accuracy.
Fixed
(49) Line 613: please change "effect in" to "effect on".
Fixed
(50) Line 614: Please clarify the number of embryos or cells and also add how many independent replicate experiments this data is representative of. Data in Figure 2 also requires a definition of P-value for ****.
Fixed
(51) Line 625: Please clarify the number of embryos or cells and also add how many independent replicate experiments this data is representative of. Data in Figure 3 also requires a definition of P-value for ****.
Fixed
(52) Line 627: description requires editing to improve accuracy "...is only slightly increased at the 8-cell stage after exposure to reversine and AZ3146". However, the results show significantly higher DNA damage with Reversine treatment, but not with AZ when compared to DMSO. Please amend accordingly.
Fixed
(53) Line 629: Please define the error bars. SEM/95% confidence interval?
Fixed
(54) Line 634-635: it is written here that chimeras were made from 1:1 DMSO/AZ3146 and Reversine/DMSO; but Figure 4A shows 1:1 DMSO(grey):AZ3146(blue), and Reversine(red):AZ3146(blue), which contradicts the legend + method section; see comments for Line 427-429. Please amend these sections accordingly.
Fixed
(55) Line 648: reversine/AZ3146 chimeras? Refer to comments above.
Fixed
(56) Line 649-650: ...AZ-treated blastomeres contribute similarly to reversine-blastomeres to the TE and EPI but significantly increase contribution to the EPI? Please add the appropriate comparison group.
Fixed
(57) Line 652: Please clarify the number of embryos or cells and also add how many independent replicate experiments this data is representative of.
Fixed
(58) Line 664: Please clarify the number of embryos or cells and also add how many independent replicate experiments this data is representative of.
Fixed
(59) Line 675-677: FigS1B legend requires a definition of P-value for * and ****, can omit **
Fixed
(60) Line 678-680: FigS1C and S1D legend: sample size and replicates? Only mentioned in Lines 117-120, which requires back calculation.
Fixed
(61) Line 682-694: (1) Fig. S2B legend: missing P-value description for *** and ****; statistical test not stated, please add. Also, Figure S2E, only requires the definition for *, and can omit others.
Fixed
(62) Line 702: FigS3B: missing description for ****, omit others.
Fixed
(63) Line 704-705: missing description for Rev/AZ group and hypoxia vs. normoxia conditions.
Fixed
(64) Line 712-713: "n>27 per treatment" Please clarify whether this refers to the number of embryos or cells and also add how many independent replicate experiments this data is representative of. Data in Figure S5 requires the definition of P-values for *, ****. Please remove for **, ***.
Fixed
(65) Line 713-715: could benefit from a description of which were marked from mTmG; e.g. why is DMSO, Rev, Rev in Green for [D]; does this mean 2-cell stage chimeras were only made with embryos treated with DMSO and Reversine? Has it been tested if you did this with AZ3146, do the proportions remain the same? This would be interesting to know.
DMSO and reversine are in green because they are the cells mark with green in the chimeras. We also did chimeras with AZ3146. Hope this clarifies.
(66) Line 719-721: why is there a difference between the proportion of aneuploid cells for the different chimeras? AZ in D/AZ, and R/AZ groups; while only R in D/R group? Is this because you only count those that were marked with mTmG (e.g. based on [Fig S5D])? (67) Line 724: low- and medium-grade chimeras would indicate quality, recommend adding low/medium grade aneuploid/mosaic chimeras.
Fixed
(68) Line 725-729: it may be my mistake, but I think the results description is not found within the Results section, but only here in the legend? Please include this detail also in the Results section.
Fixed
(69) Line 729: which is AZ or Rev cells?
(70) References - Page number missing for some references; abbreviated version vs. non abbreviated version of journal titles used. Please be consistent/meet journal requirements.
Fixed
(71) Figures
Figure 1: [C] both AZ-NANOG and DMSO-SOX17 have mean/median(?) of 11 cells (described in results), yet in this figure (on the same axis) these groups are not level. Are the numbers correct? This is also the case for Rev-SOX17 which is described in the results as having 8 cells yet appears to be above the 8 mark in the graphs; AZ-CDX2, which has 64 cells yet appears to be below the 60 mark; AZ-total, which has 82 cells yet appears to be below the 80 mark. In [E] the label orientation, "ns" has both horizontal and vertical orientation. Please make appropriate changes throughout to reflect accuracy.
Figure 3: [C] As for Figure 1, DMSO-NANOG, which is described in results as having 14 cells, yet appears to be below the 13 mark in the graph; DMSO-SOX17, which has 6 cells yet appears to be above the 7 mark.
These is due to average
Figure 4: [D and E] random numerals appear in the bars on the graph. 9,10 and 7, 14? Are these sample size numbers? If they are, they should appear in all bars/groups or in the legend.
Yes, these are sample sizes
Figure 5: [D and G] same comment as for Fig 4 above, random numbers in the graph.
Yes, these are sample sizes
(72) Supplementary figures. Figure S2 [A] No quantification? This is important to add as representative images are only a 2D plane, which can be easily misinterpreted. [E] Should the y-axis label be written as "Number of cells normalised to DMSO group", or similar? Or is there a figure missing to depict the ratio of cells in each cell lineage normalised to the DMSO group, which is the description written in the legend? But I don't see a figure showing the ratio, just the absolute number of cells. Is this a missing figure or a mislabelled axis?
Quantification at the blastocyst stage is misleading due to high cellular heterogeneity.
Reviewer #3 (Recommendations for the authors):
(1) The statement in the abstract: "embryos with a low proportion of aneuploid cells have a similar likelihood of developing to term as fully euploid embryos" Line 48-50 Capalbo does not really answer as the biopsy may not be reflective of ICM.
This is a great point. Trophectoderm biopsies may not reflect the real proportion of aneuploidy in the ICM. We emphasize this in discussion and Fig. S4.
(2) Line 69/70, at least 50% Singla et al/Bolton. It would be helpful to elaborate a bit more on this study. How can this be assessed when analysis results in destruction?
(3) Differences in the developmental potential of reversine versus AZ-treated embryos. It is not entirely clear why. The differences in non-dividing cells if any are small, and the -crest cells are rather minor also. Could these drugs have other effects that are not evaluated in the study?
Yes, specifically, reversine has been shown to have several off-targets effects. Including inducing apoptosis (Chen et al 2024).
(4) Lines 45-46 understanding of reduction of aneuploidy should mention/discuss the paper of attrition/selection, of the kind by the Brivanlou lab for instance, or others. As well as allocation to specific lineages, including the authors' work.
Dr. Brinvanlou experiments in gastruloids do not represent the same developmental stage of pre-implantation embryos. Comparison between models is debatable.
(5) Line 53: human experiments are more limited due to access to samples. What does 'not allowed' mean? By who, where?
NIH does not allow to experiment with human embryos for ethical reasons.
(6) The figure callouts to S1A in lines 93,97. What is a non-dividing nucleus? For how long is it observed?
A non-dividing nucleus is an accumulation of DNA in a round form without define separation of the chromosomes and their specific kinetochores (CREST antibody). The presence of non-dividing nucleus during the 4 -to-8 cell stage can indicate activation of the spindle assembly checkpoint during prometaphase. Example of non-dividing nucleus can be observed in Fig S1.B.
(7) Line 108 A relatively minor effect on cell number and quality of blastocysts is observed. It is not surprising that thereafter, developmental potential is also high. At that stage, what are the individual cell karyotypes?
Due to technical limitations, we can’t determine the specific karyotypes of these cells.
(8) Line 153. The p53 increase of 1.3 fold is not dramatic.
The levels of p53 at the morula stage is 7-fold differences. In contrast, at the blastocyst stage, a change in 1.3-fold is indeed less dramatic. This can be a result of the elimination of aneuploid cells or mechanism to counter the activation of the p53 pathway, like overexpression of the Hif1a pathway.
(9) Line 155. Is there a more direct way to test for p38 activation?
Natale et al 2004 (Dev Biol) and Sozen et al 2015 (Mech of Dev) described that inhibition of p38 deeply affect the development of pre-implantation embryos after the 8-cell stage. For this reason, comprehensible dissect the interaction between p53, HIF1A and p38 during aneuploid stress is challenging. We do not discard a double function of p38 during lineage specification and in response to DNA damage.
(10) Line 191/192 Low oxygen conditions, is this equal to hypoxia? What is the definition of hypoxia here? The next sentence says physiological. Is that the same or different?
Low oxygen can be defined as hypoxia. This varies from 2% to 6%. Our definition of hypoxia is 5% concentration of oxygen with 5% concentration of CO2, taking into consideration the standard levels of oxygen in the IVF clinics. Physiological oxygen in mouse varies from ~1.5% to 8%.
(11) The question is whether there is something specific about HIF1 and aneuploidy, or whether another added stress would have similar effects on the competitiveness of treated cells.
That is a great follow up of our work.
(12) Line 300. Is p21 unregulated at the protein level or mRNA level? Please indicate.
mRNA level.
(13) Figure 1D/E H2Ax intensity is cell cycle phase-dependent. It might be meaningful to count foci by the nucleus and show both ways of analysis.
(14) Check the spelling of phalloidin.
Fixed in text and figures!
-

eLife Assessment
Sanchez-Vasquez et al establish an innovative approach to induce aneuploidy in preimplantation embryos. This important study extends the author's previous publications evaluating the consequences of aneuploidy in the mammalian embryo. In this work, the authors investigate the developmental potential of aneuploid embryos and characterize changes in gene expression profiles under normoxic and hypoxic culture conditions. Using a solid methodology they identify sensitivity to Hif1alpha loss in aneuploid embryos, and in further convincing experiments they assess how levels of DNA damage and DNA repair are altered under hypoxic and normoxic conditions.
-

Reviewer #1 (Public review):
Summary:
This paper developed a model of chromosome mosaicism by using a new aneuploidy-inducing drug (AZ3146), and compared this to their previous work where they used reversine, to demonstrate the fate of aneuploid cells during murine preimplantation embryo development. They found that AZ3146 acts similarly to reversine in inducing aneuploidy in embryos, but interestingly showed that the developmental potential of embryos is higher in AZ3146-treated vs. reversine-treated embryos. This difference was associated with changes in HIF1A, p53 gene regulation, DNA damage, and fate of euploid and aneuploid cells when embryos were cultured in a hypoxic environment.
Strengths:
In the current study, the authors investigate the fate of aneuploid cells in the preimplantation murine embryo using a specific …
Reviewer #1 (Public review):
Summary:
This paper developed a model of chromosome mosaicism by using a new aneuploidy-inducing drug (AZ3146), and compared this to their previous work where they used reversine, to demonstrate the fate of aneuploid cells during murine preimplantation embryo development. They found that AZ3146 acts similarly to reversine in inducing aneuploidy in embryos, but interestingly showed that the developmental potential of embryos is higher in AZ3146-treated vs. reversine-treated embryos. This difference was associated with changes in HIF1A, p53 gene regulation, DNA damage, and fate of euploid and aneuploid cells when embryos were cultured in a hypoxic environment.
Strengths:
In the current study, the authors investigate the fate of aneuploid cells in the preimplantation murine embryo using a specific aneuploidy-inducing compound to generate embryos that were chimeras of euploid and aneuploid cells. The strength of the work is that they investigate the developmental potential and changes in gene expression profiles under normoxic and hypoxic culture conditions. Further, they also assessed how levels of DNA damage and DNA repair are altered in these culture conditions. They also assessed the allocation of aneuploid cells to the divergent cell lineages of the blastocyst stage embryo.
Weaknesses:
Inconsistent/missing description for sample size, biological/technical replicates, label orientation, the appropriate number of * for each figure panel, and statistical tests used.
-

Reviewer #2 (Public review):
Summary:
This study by Sanchez-Vasquez is a very innovative approach to inducing aneuploidy and then studying the contribution of treated cells to different lineages, including post-implantation. It connects well to the authors' previous work to induce mosaic aneuploidies. The authors identify sensitivity to HIF1a loss in treated embryos with likely aneuploidy. This work is part of an important line of work with evaluates the consequences of aneuploidy in the mammalian embryo.
Weaknesses:
Given that this is a study on the induction of aneuploidy, it would be meaningful to assess aneuploidy immediately after induction, and then again before implantation. This is also applicable to the competition experiments on page 7/8. What is shown is the competitiveness of treated cells. Because the publication centers …
Reviewer #2 (Public review):
Summary:
This study by Sanchez-Vasquez is a very innovative approach to inducing aneuploidy and then studying the contribution of treated cells to different lineages, including post-implantation. It connects well to the authors' previous work to induce mosaic aneuploidies. The authors identify sensitivity to HIF1a loss in treated embryos with likely aneuploidy. This work is part of an important line of work with evaluates the consequences of aneuploidy in the mammalian embryo.
Weaknesses:
Given that this is a study on the induction of aneuploidy, it would be meaningful to assess aneuploidy immediately after induction, and then again before implantation. This is also applicable to the competition experiments on page 7/8. What is shown is the competitiveness of treated cells. Because the publication centers around aneuploidy, the inclusion of such data in the main figure at all relevant points would strengthen it. There is some evaluation of karyotypes only in the supplemental - why? It would be good not to rely on a single assay that the authors appear to not give much importance.
-

-

Excerpt
Hypoxia regulates embryonic fitness in mosaic aneuploid embryos
-