Dysregulated H19/Igf2 expression disrupts cardiac-placental axis during development of Silver-Russell syndrome-like mouse models
Curation statements for this article:-
Curated by eLife
eLife assessment
Igf2 and H19 are the two best-studied imprinted genes in mice. Taking advantage of the varying levels of H19 and Igf2 expression in three existing mouse models, the authors dissect the role of H19 and Igf2 in cardiac and placental development. Their findings suggest that an accurate dosage of both H19 and Igf2 is critical for normal embryonic development, especially the development of the heart and placenta. The work is of interest to colleagues studying imprinting as well as mammalian development.
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Dysregulation of the imprinted H19/IGF2 locus can lead to Silver-Russell syndrome (SRS) in humans. However, the mechanism of how abnormal H19/IGF2 expression contributes to various SRS phenotypes remains unclear, largely due to incomplete understanding of the developmental functions of these two genes. We previously generated a mouse model with humanized H19/IGF2 imprinting control region ( hIC1 ) on the paternal allele that exhibited H19/Igf2 dysregulation together with SRS-like growth restriction and perinatal lethality. Here, we dissect the role of H19 and Igf2 in cardiac and placental development utilizing multiple mouse models with varying levels of H19 and Igf2 . We report severe cardiac defects such as ventricular septal defects and thinned myocardium, placental anomalies including thrombosis and vascular malformations, together with growth restriction in mouse embryos that correlated with the extent of H19/Igf2 dysregulation. Transcriptomic analysis using cardiac endothelial cells of these mouse models shows that H19/Igf2 dysregulation disrupts pathways related to extracellular matrix and proliferation of endothelial cells. Our work links the heart and placenta through regulation by H19 and Igf2 , demonstrating that accurate dosage of both H19 and Igf2 is critical for normal embryonic development, especially related to the cardiac-placental axis.
Article activity feed
-

-

Author Response
Reviewer #2 (Public Review):
Point 1: The transcriptomic analysis of E12.5 endocardial cushion cells in the various mouse models is informative in the extraction of Igf2- and H19-specific gene functions. In Fig. 6D, a huge sex effect is obvious with many more DEGs in female embryos compared to males. How can this be explained given that Igf2/H19 reside on Chr7 and do not primarily affect gene expression on the X chromosome? Is any chromosomal bias observed in the genomic distribution of DEGs?
We examined chromosomal distribution of DEGs between WT and +/hIC1 (Supplemental Figure 6D) and did not see any bias on X chromosome. We described this result on lines 278-280: “Although the number of +/hIC1-specific DEGs largely differed between males and females, there was no sex-specific bias on the X chromosome (Supplemental …
Author Response
Reviewer #2 (Public Review):
Point 1: The transcriptomic analysis of E12.5 endocardial cushion cells in the various mouse models is informative in the extraction of Igf2- and H19-specific gene functions. In Fig. 6D, a huge sex effect is obvious with many more DEGs in female embryos compared to males. How can this be explained given that Igf2/H19 reside on Chr7 and do not primarily affect gene expression on the X chromosome? Is any chromosomal bias observed in the genomic distribution of DEGs?
We examined chromosomal distribution of DEGs between WT and +/hIC1 (Supplemental Figure 6D) and did not see any bias on X chromosome. We described this result on lines 278-280: “Although the number of +/hIC1-specific DEGs largely differed between males and females, there was no sex-specific bias on the X chromosome (Supplemental Figure 6D).” Additionally, we agree with the reviewer that it is noteworthy that the dysregulated H19/Igf2 expression affected transcriptome in a sex-specific manner, especially when the mutation is located on a somatic chromosome. Although investigating the role of hormones versus sex chromosome in these effects would be quite interesting, it is beyond the scope of current study.
Point 2: A separate issue is raised by Fig. 6E that shows a most dramatic dysregulation of a single gene in the delta3.8/hIC1 "rescue" model. Interestingly, this gene is Shh. Hence, these embryos should exhibit some dramatic skeletal abnormalities or other defects linked to sonic hedgehog function.
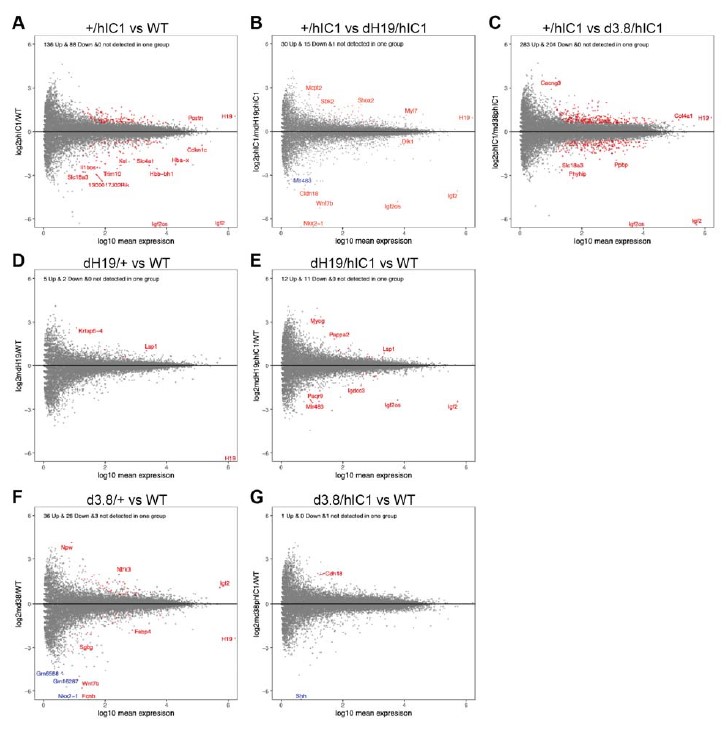
The reason why Shh appeared to be differentially expressed between wild-type and d3.8/hIC1 samples was that Shh expression was 0 across all the samples except for two wild-type samples. In order to detect all the DEGs that might be lowly expressed, we did not want to filter DEGs based on the level of total expression. As a result, Shh was represented as significantly differently expressed in d3.8/hIC1 samples, although its expression in our samples appears to be too low to have any significant effect on development. This explanation was added to lines 310-312. To confirm that this was an exceptional case, we analyzed the expression of DEGs obtained from other pairwise comparisons. In the volcano plots below, genes of which expression is not statistically different between two groups are marked grey. Genes of which expression is statistically different and detected in both groups are marked red. Genes with statistically different but not detected in one group at all, such as Shh, are marked blue (Figure G). It is clear that that almost all of our DEGs are expressed consistently across the groups, and genes with no expression detected in one group are very rare.
Point 3: The placental analysis needs to be strengthened. Placentas should be consistently positioned with the decidua facing up, and the chorionic plate down. The placentas in Fig. 3F are sectioned at an angle and the chorionic plate is missing. These images must be replaced with better histological sections.
As requested, we have replaced placental images with better representative sections (Figure 3F and 4E). In addition, we have improved alignment of placental histology figures.
Point 4: The CD34 staining has not worked and does not show any fetal vasculature, in particular not in the WT sample.
As requested, we have replaced the CD34 vascular stained images with those that better represent fetal vasculature (Figure 3G).
Point 5: The "thrombi" highlighted in Fig. 4E are well within the normal range, to make the point that these are persistent abnormalities more thorough measurements would need to be performed (number, size, etc).
As requested, we measured the number and relative size of the thrombi that are found in dH19/hIC1 placentas with lesions. No thrombi were found in wild-type placentas whereas an average of 1.3 thrombi were found in six dH19/hIC1 placentas. The size of the thrombi widely varied, but occupied average of 2.58% of the labyrinth zone where these lesions were found (Supplemental Figure 4D). Additionally, we replaced the image in Figure 4E into the section that better represents the lesion.
Point 6: The statement that H19 is disproportionately contributing to the labyrinth phenotype (lines 154/155) is not warranted as Igf2 expression is reduced to virtually nothing in these mice. Even though there is more H19 in the labyrinth than in the junctional zone, the phenotype may still be driven by a loss of Igf2. Given the quasi Igf2-null situation in +/hIC1 mice, is the glycogen cell type phenotype recapitulated in these mice, and how do glycogen numbers compare in the other mouse models?
The sentence was edited in line 157. We performed Periodic acid Schiff (PAS) staining on +/hIC1 placentas to address if glycogen cells are affected by abnormal H19/Igf2 expression (Supplemental Figure 1E). In contrary to previous reports where Igf2-null mice had lower placental glycogen concentration (Lopez et al., 1996) and H19 deletion led to increased placental glycogen storage (Esquiliano et al., 2009), our quantification on PAS-stained images showed that the glycogen content is not significantly different between wild-type and +/hIC1 placentas. We have described this result in lines 166-168.
Point 7: How do delta3.8/+ and delta3.8/hIC1 mice with a VSD survive? Is it resolved some time after birth such that heart function is compatible with postnatal viability? And more importantly, do H19 expression levels correlate with phenotype severity on an individual basis?
Our study was limited to phenotypes prior to birth, thus postnatal/adult phenotypes were not examined. Because the VSD showed only partial penetrance in these mice, we cannot state that the d3.8/+ or d3.8/hlC1 mice with VSDs survive. It has also been previously reported in another mouse model with incomplete penetrance of a VSD that the mice which survived to adulthood did not have the VSDs (Sakata et al., 2002). We find it highly unlikely that either mouse model would survive significantly past the postnatal timepoint with a VSD. We have examined two PN0 d3.8/hIC1 neonates, and both did not have VSD.
Regarding the second point, the only way to quantitatively address this question would be to do qPCR or RNA-seq on individual hearts, which then makes it impossible for those hearts to be examined for histology to confirm the VSD. Thus, hearts used to identify VSDs via histology could not also be used for quantitative H19 measurements. One thing to note is that the H19/Igf2 expression in independent replicates of d3.8/hIC1 cardiac ECs used in our RNA-seq experiment is quite variable, not clustering together in contrast to other mouse models used in this study (Fig. 6A). Such wide range of variability in the extent of H19/Igf2 dysregulation suggests that H19/Igf2 levels could have an impact on the penetrance or the severity of the VSD phenotype in d3.8/hIC1 embryos.
-

eLife assessment
Igf2 and H19 are the two best-studied imprinted genes in mice. Taking advantage of the varying levels of H19 and Igf2 expression in three existing mouse models, the authors dissect the role of H19 and Igf2 in cardiac and placental development. Their findings suggest that an accurate dosage of both H19 and Igf2 is critical for normal embryonic development, especially the development of the heart and placenta. The work is of interest to colleagues studying imprinting as well as mammalian development.
-

Reviewer #1 (Public Review):
Using multiple mouse models with varying levels of H19 and Igf2 expression, the authors dissect the role of H19 and Igf2 in cardiac and placental development. Severe cardiac defects and placental anomalies were found to be correlated with the extent of H19/Igf2 dysregulation. Transcriptomic analysis revealed that H19/Igf2 dysregulation disrupts pathways related to extracellular matrix (ECM) and proliferation of endothelial cells. This work links the heart and placenta through regulation by H19 and Igf2, demonstrating that an accurate dosage of both H19 and Igf2 is critical for normal embryonic development, especially related to the cardiac-placental axis. The topic is of significance and the data are of high quality.
-

Reviewer #2 (Public Review):
Despite the fact that Igf2 and H19 are the two best-studied imprinted genes in (mouse) development, substantial gaps in knowledge continue to exist as to their independent functions in normal and pathological situations, including in the imprinting disorders BWS and SRS to which they are linked. Here, using three established mouse models in a clever way, the authors are able to dissect the impact of overexpression or depletion of Igf2 and H19 independently of each other. This sophisticated use of mouse genetic models is a major achievement in trying to discern the precise impact of these genes on the development of particular cell types and tissues.
The authors report the placenta and heart as the two most severely affected organs in these mouse models. What remains unclear is if these phenotypes are …
Reviewer #2 (Public Review):
Despite the fact that Igf2 and H19 are the two best-studied imprinted genes in (mouse) development, substantial gaps in knowledge continue to exist as to their independent functions in normal and pathological situations, including in the imprinting disorders BWS and SRS to which they are linked. Here, using three established mouse models in a clever way, the authors are able to dissect the impact of overexpression or depletion of Igf2 and H19 independently of each other. This sophisticated use of mouse genetic models is a major achievement in trying to discern the precise impact of these genes on the development of particular cell types and tissues.
The authors report the placenta and heart as the two most severely affected organs in these mouse models. What remains unclear is if these phenotypes are causally linked, such that - for example - endothelial cell dysfunction causes the placental phenotypes or, conversely, trophoblast dysfunction causes the heart phenotypes. The "placenta-heart axis" is repeatedly referred to; however, unlike in the use of the term in the current manuscript, this term is more commonly used to describe situations where the placenta is causative of heart phenotypes, as first established in the Pparg mutation. Along these lines, it would be instrumental to establish in at least one of the mouse models under investigation here, if any of the heart or placental defects are secondary to gene function in a cell type outside of the affected organ itself.
The transcriptomic analysis of E12.5 endocardial cushion cells in the various mouse models is informative in the extraction of Igf2- and H19-specific gene functions. This analysis raises a few questions: In Fig. 6D, a huge sex effect is obvious with many more DEGs in female embryos compared to males. How can this be explained given that Igf2/H19 reside on Chr7 and do not primarily affect gene expression on the X chromosome? Is any chromosomal bias observed in the genomic distribution of DEGs? A separate issue is raised by Fig. 6E that shows a most dramatic dysregulation of a single gene in the delta3.8/hIC1 "rescue" model. Interestingly, this gene is Shh. Hence, these embryos should exhibit some dramatic skeletal abnormalities or other defects linked to sonic hedgehog function.
The placental analysis needs to be strengthened. Placentas should be consistently positioned with the decidua facing up, and the chorionic plate down. The placentas in Fig. 3F are sectioned at an angle and the chorionic plate is missing. These images must be replaced with better histological sections. The CD34 staining has not worked and does not show any fetal vasculature, in particular not in the WT sample. The "thrombi" highlighted in Fig. 4E are well within the normal range, to make the point that these are persistent abnormalities more thorough measurements would need to be performed (number, size, etc).
The statement that H19 is disproportionately contributing to the labyrinth phenotype (lines 154/155) is not warranted as Igf2 expression is reduced to virtually nothing in these mice. Even though there is more H19 in the labyrinth than in the junctional zone, the phenotype may still be driven by a loss of Igf2.
Given the quasi Igf2-null situation in +/hIC1 mice, is the glycogen cell type phenotype recapitulated in these mice, and how do glycogen numbers compare in the other mouse models?How do delta3.8/+ and delta3.8/hIC1 mice with a VSD survive? Is it resolved some time after birth such that heart function is compatible with postnatal viability? And more importantly, do H19 expression levels correlate with phenotype severity on an individual basis?
-

Reviewer #3 (Public Review):
The study conducted by Chang and colleagues elegantly describes the significance of appropriate H19 and Igf2 gene expression control in the formation of the fetal heart and placenta. They used established and newly developed genetic models in mice, histological analyses, and transcriptomic assessments to assess the contribution of H19 and Igf2 to the defects observed. On a whole the paper is very well written, the experimental design is sound, the results compelling, and the conclusions supported. I only have minor suggested edits/comments.
-