Transcriptome network analysis implicates CX3CR1-positive type 3 dendritic cells in non-infectious uveitis
Curation statements for this article:-
Curated by eLife
Evaluation Summary:
The authors provide evidence suggesting the gene expression profile of a specific subset of dendritic cells define features of specific forms of non-infectious uveitis. This work suggests specific pathways in these cells that may have mechanistic import in inflammatory eye disease. This manuscript is of interested to immunologists studying autoimmunity and ocular immunity. While the paper is largely descriptive, the data it presents should serve as a valuable resource for generating hypotheses about the pathogenesis of ocular autoimmune disease.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #1 agreed to share their name with the authors.)
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Type I interferons (IFNs) promote the expansion of subsets of CD1c+ conventional dendritic cells (CD1c+ DCs), but the molecular basis of CD1c+ DCs involvement in conditions not associated without elevated type I IFNs remains unclear.
Methods:
We analyzed CD1c+ DCs from two cohorts of non-infectious uveitis patients and healthy donors using RNA-sequencing followed by high-dimensional flow cytometry to characterize the CD1c+ DC populations.
Results:
We report that the CD1c+ DCs pool from patients with non-infectious uveitis is skewed toward a gene module with the chemokine receptor CX3CR1 as the key hub gene. We confirmed these results in an independent case–control cohort and show that the disease-associated gene module is not mediated by type I IFNs. An analysis of peripheral blood using flow cytometry revealed that CX3CR1+ DC3s were diminished, whereas CX3CR1− DC3s were not. Stimulated CX3CR1+ DC3s secrete high levels of inflammatory cytokines, including TNF-alpha, and CX3CR1+ DC3 like cells can be detected in inflamed eyes of patients.
Conclusions:
These results show that CX3CR1+ DC3s are implicated in non-infectious uveitis and can secrete proinflammatory mediators implicated in its pathophysiology.
Funding:
The presented work is supported by UitZicht (project number #2014-4, #2019-10, and #2021-4). The funders had no role in the design, execution, interpretation, or writing of the study.
Article activity feed
-

-

Author Response
Reviewer #1 (Public Review):
In the article "Whole transcriptome-sequencing and network analysis of CD1c+ human dendritic cells identifies cytokine-secreting subsets linked to type I IFN-negative autoimmunity to the eye," Hiddingh, Pandit, Verhagen, et al., analyze peripheral antigen presenting cells from patients with active uveitis and control patients, and find several differentially expressed transcription factors and surface markers. In addition, they find a subset of antigen presenting cells that is decreased in frequency in patients with uveitis that in previous publications was shown to be increased in the eye of patients with active uveitis. The greatest strength of this paper is the ability to obtain such a large number of samples from active uveitis patients that are not currently on systemic therapy. While …
Author Response
Reviewer #1 (Public Review):
In the article "Whole transcriptome-sequencing and network analysis of CD1c+ human dendritic cells identifies cytokine-secreting subsets linked to type I IFN-negative autoimmunity to the eye," Hiddingh, Pandit, Verhagen, et al., analyze peripheral antigen presenting cells from patients with active uveitis and control patients, and find several differentially expressed transcription factors and surface markers. In addition, they find a subset of antigen presenting cells that is decreased in frequency in patients with uveitis that in previous publications was shown to be increased in the eye of patients with active uveitis. The greatest strength of this paper is the ability to obtain such a large number of samples from active uveitis patients that are not currently on systemic therapy. While the validation experiments have methodologic flaws that decrease their usefulness, this study will still serve as a valuable resource in generating hypotheses about the pathogenesis of uveitis that can be tested in future projects.
We thank the reviewer for the constructive comments and effort to review our work in detail.
Since all CD36+CX3CR1+ cells are CD14+ (Figure 4D), how CX3CR1 ended up being differentially regulated in a similar way despite this population was excluded from 2nd bulk RNAseq data set should be commented on by the authors.
We agree with reviewer that the CD14 surface expression in relation to the black-gene module and CD36+CX3CR1+ DC3s requires more detailed analysis. As described in the results section, genes in this module are linked to both CD1c+ DCs and inflammatory CD14+ monocytes, which we cannot distinguish by bulk RNA seq analysis. Therefore, we aimed to use an approach to demonstrate that the black module is a bona fide CD1c+ DC gene signature not dependent on CD14 surface expression: We showed that there was not difference in CD14+ cell fractions in the samples for RNA-seq between patient and control samples (see Fig. 1F). We now further investigated this by additional data and experiments. We now show in Figure 2 Supplement 2A that CD14 – as expected - does not correlate with the black module. To confirm this experimentally, we purified CD14+CD1c+ and CD14- CD1c+ DCs from 6 donors and subjected these to qPCR analysis to evaluate the expression of key genes from the black module (see revised Figure 2A). As illustrated in revised Figure 2 panel B, we show that the expression levels of genes, including CD36 and CX3CR1, are not significantly altered between CD14+/- CD1c+ DCs which supports that the identified gene module is also not dependent on CD14 surface expression by CD1c+ DCs. To assess if the expression of the black module was also independent of CD14 in inflammatory disease, we used RNA-seq data from FACS-sorted CD14+CD1c+ DCs and CD14-CD1c+ DCS from patients with SLE and Scleroderma (GSE13731) and confirm that the expression of the black module genes is independent from CD14 surface expression (see revised Figure 2 panel C). Finally, we removed CD14+ cells from the analysis in the 2nd bulk RNA-seq experiment to proof that indeed the black module could be perceived as being associated with uveitis independent of CD14+ expression which allowed attributing the black module to CD1c+ DCs by bulk RNA-seq analyses. Also, more detailed analysis by flow cytometry (Revised Figure 4) and scRNA-seq (Figure 6) confirm these findings. For example, we show that the CD36+ CX3CR1+ DC3s are in fact a subset of CD14+ CD1c+ DCs (Figure 2 – Supplement 2) and we show that eye-infiltrating CD1c+ DCs that harbor the black module gene signature show increased CD36 and CX3CR1, but not CD14 (Figure 6C). We have addressed all these experiments and data in the result section on page 12-13, 16,17, and in the discussion section on page 19. We hope the reviewer agrees that this has now been sufficiently addressed.
Line 153: "...substantiates this gene set as a core transcriptional feature of human autoimmune uveitis." It would be difficult to argue that when only 137 of the 1236 DEGs from the first module are repeated in a validation data set that this is the core transcriptions set that defines the population in any uveitis. Further concerns include that the validation data set is not the same population, but rather a subset not containing CD14.
We agree with the reviewer and have changed this in the result section to “substantiates this gene set as a robust and bona fide transcriptional feature of CD1c+ DCs in human non-infectious uveitis” at page 13. We agree that - as expected - the removal of CD14+ cells impacted the sensitivity of our analysis, but that this strategy was required to attribute the black module to CD1c+ DCs. Our data supports that the black module gene signature is not restricted to CD14+ CD1c+ DCs by demonstrating that its dysregulation in non-infectious uveitis can even be perceived in CD14- CD1c+ DCs. We show now that the replication of a fraction of genes of the black module is a consequence of sensitivity to detect differentially expressed genes (Figure 2 – Supplement 1C). – most likely due to lower cell number after sorting out CD14+ cells. We have outlined this in greater detail in the result section on page 13. We hope the reviewer agrees this has now been adequately described.
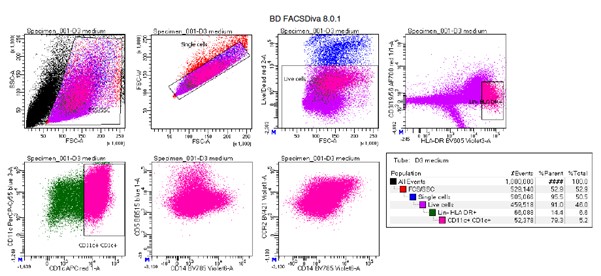
Line 220: Notch-dll experiments: with the experiments presented it is not possible to say that the changes are due to maintenance of CD1c+ DCs without further experiments outlining what NOTCH2 signaling changes throughout time. Is the population fully developed in the first 7 days of culture prior to adding NOTCH2 or ADAM10 inhibitors? Is there more apoptosis in this pathway? Less proliferation? It would be more accurate to say that there are fewer cDC2s after 14 days of culture without speculating the cause. In this experiment it is unclear why the gate of CD141/CD1c was chosen, as this appears to be in the middle of the population. In normal PBMCs CD141+ DCs would be CD1c negative; therefore why exclude the CD141hiCD1c+ and CD141loCD1c+ populations?
We agree with the reviewer that in the current state the additional Notch-DLL experiments are inconclusive. Based on the comments from this reviewer, we believe the most appropriate experiments would be to show changes in the surface protein expression of CD36, CX3CR1 and other key surface markers of the black module upon inhibition of NOTCH2 or ADAM10. To this end, we repeated the experiments with human CD34+ HPC-derived DCs cells to measure cell subset by flow cytometry using the same panel we used for the PBMCs. However, we experienced substantial autofluorescence of human CD34-HPC derived cultures (expected for the complex heterogeneous cellularity of these cultures and as previously reported for CD34+ cells (Donnenberg et al., Methods 2015) that introduced significant artifacts and interfere with optimal identification of CD1c+ DCs and their subsets (see example below). We were unable to control for this so far, unfortunately. Since we agree with the reviewer that in the current form the supplemental figure does not significantly contribute to the manuscript, we removed the supplemental figure entirely from the manuscript. We hope the reviewer agrees that we already provide several complementary lines of evidence that link NOTCH-RUNX3 signaling to the black module (Figure 3A-D), including RNA-seq data from NOTCH2-DLL experiments, and that the current data is sufficient to support the main conclusions of the manuscript. We hope the reviewer agrees with this proposal.
Author response figure 1: Manual gating example of human CD34-HPC derived DCs shows substantial autofluorescence.
Line 256: The hypothesis that the loss of CD36+CX3CR1+ cells was due to migration to the eye doesn't make sense based on volume and number of cells. 0.1% of all PBMC is ~1x107 cells, and distributed throughout the eye would give about 1.3x106 cells/mL of eye volume. This would make the eye turbid which is not consistent with birdshot chorioretinopathy and would be rare in HLA-B27 anterior uveitis and intermediate uveitis
We agree with the reviewer and have changed this in the manuscript section to “We speculated that the decrease in blood CD36+CX3CR1+ CD1c+ DCs was in part the result of migration of these cells to peripheral tissues (lymph nodes) and that these cells may also infiltrate the eye during active uveitis.” On page 17.
Line 267: Would have liked to see the gating of CX3CR1/CD36 cells be more consistent (there are overlapping CX3CR1+ and CX3CR1- populations in 5A, but in Figure 4 quadrants were used to define the populations when evaluating the numbers in uveitis and healthy controls. The populations in Figure 5 are more separated by CD36.
We agree with the reviewer and have added a more detailed example of the gating strategy used to sort CD36/CX3CR1 subsets in Figure 5 – Supplement 1 including the expression of CX3CR1 and CD36 in the sorted populations.
Line 269, IN VITRO stimulation: The experimental paradigm is set up to find a difference between cells but does not to test any biologically relevant scenario. By sorting on a surface marker, then stimulating with the ligand for that receptor, the result better proves that CD36 is important in TLR2 signaling than does it give any information on how these dendritic cells might behave in uveitis.
We agree with the reviewer that the connection between the cytokine expression of the CD1c+ subsets and non-infectious uveitis may benefit from additional experimental data. To this end, we profiled available eye fluid biopsies and paired plasma by Olink proteomics to measure 92 immune mediators from patients and controls from this study (and several additional samples, including aqueous humor from non-inflammatory cataract controls – see revised Figure 5 panel D). This analysis shows that cytokines produced by CD36+CX3CR1+ DCs such as TNF-alpha and IL-6 are specifically increased in eye tissue of patients, but not in blood. We hope the reviewer agrees that we have provided additional experimental data that links the functional differences in DC subsets to cytokines implicated in the pathogenesis of non-infectious uveitis.
Reviewer #3 (Public Review):
First, a note on nomenclature. The authors use the term 'auto-immune' uveitis to encapsulate three different conditions -- HLA-B27 anterior uveitis, idiopathic intermediate uveitis, and birdshot choroidopathy. While I would agree with this terminology for the third set, there is substantial controversy as to whether HLA-B27 is truly autoimmune or autoinflammatory. Indeed, one major hypothesis is that this condition is driven by changes in gut microbiome. Intermediate uveitis is even more problematic; a substantial number of cases of this condition will turn out to be associated with demyelinating disease, which has recently been linked to Epstein Barr virus disease. To my knowledge in none of these diseases has a definitive autoantigen been identified nor passive transfer via transfusion shown; I would suggest the authors abandon this terminology and simply refer to the conditions as they are called.
We would like to thank the reviewer for the constructive suggestions. We agree and have changed the term “autoimmune uveitis” to “non-infectious uveitis” throughout the manuscript.
Further, it would have been very desirable to compare the DC transcriptome for the other class of uveitic disease -- infectious -- for acute retinal necrosis or similar. As well it would have been very useful to compare profiles to other, related immune-mediated diseases such as ankylosing spondylitis.
We agree with the reviewer that comparison of DC transcriptomes is useful for interpretation of biological mechanisms involved. This is precisely the reason we use (in Figure 3) comparison of our DC transcriptomic data to well-controlled transgenic models and DC culture systems. This revealed NOTCH2-RUNX3 signaling driving the uveitis-associated CD1c+ DC signature. We have now included transcriptomic data from CD1c+ DC subsets of type I IFN diseases SLE and Systemic Sclerosis in Figure 2. Although we agree that comparison to infectious uveitis would be interesting, bulk RNA-seq data from CD1c+ DCs are – to the best of our knowledge – unfortunately not available.
Finally, it must be noted that looking for systemic signals in dendritic gene expression may be a bit of a needle in the haystack approach. Presumably, the function of the dendritic cells in uveitis is largely centered on those cells in the eye. It would have been highly desirable to examine the expression profile of intraocular DCs in at least a subset of patients who may have come to surgery (for instance, steroid implantation or vitrectomy).
We agree with the reviewer that analysis of blood requires enormous efforts and controls to dissect disease-relevant changes in gene profiles of cDC2 subsets. We therefore designed a strategy that focusses on replication of gene modules, use independent cohorts, and complementary immunophenotyping technologies to detect key changes in specific subsets of CD1c+ DCs in uveitis patients. To further extend these analyses, we have now also detailed our analysis of intraocular DCs using single-cell RNA seq of eye fluid biopsies (aqueous humor) of HLA-B27 anterior uveitis (identical to our “AU” group of patients). As shown in revised Figure 6, we detected eye-infiltrating CD1c+ DCs and were able to cluster cells positive for the uveitis-associated black module (revised Figure 6B), which showed – as expected - that “black-module+” CD1c+ DCs show higher expression for CD36, CX3CR1, and lower RUNX3, but not CD14 (revised Figure 6C)– closely corroborating our blood CD1c+ DC analyses. These DC3s were also found at higher frequency in the eye of patients with AU (Figure 6D). We hope the reviewer agrees we have sustainably improved the analysis of intraocular DCs and that this has now been sufficiently addressed.
It is also problematic that no effort has been made to assess the severity of uveitis. Flares of disease can range from extremely mild to debilitating. Similarly, intermediate uveitis and BSCR can range greatly in severity. Without normalizing for disease severity it is difficult to fully understand the range of transcriptional changes between cases.
In our view, a key limitation in determination of uveitis severity for molecular analysis is the fact that objective biomarkers that assess disease severity across uveitis entities are lacking. Currently, disease severity is dependent an array of clinical features (i.e, SUN criteria) which cannot be applied consistently to anterior, intermediate and posterior uveitis. For example, the severity of anterior uveitis is in part assessed by grading of inflammation in the anterior chamber, while the anterior chamber is (typically) not involved in Birdshot Uveitis (BU in this study). However, to allow the study of patients with high disease activity, we exclusively used systemic treatment-free patients that all had active uveitis at sampling at our academic institute, making the results highly relevant for understanding the pathophysiology of non-infectious uveitis. For this reviewer’s convenience, we have conducted additional analysis that includes key clinical parameters (anterior chamber cells, vitreous cells, and macular thickness for patients from cohort I). These data showed no clear clustering of patients based on any of the clinical parameters (revised Figure 1 -Supplement 2). We hope the reviewer agrees this has been addressed in sufficient detail.
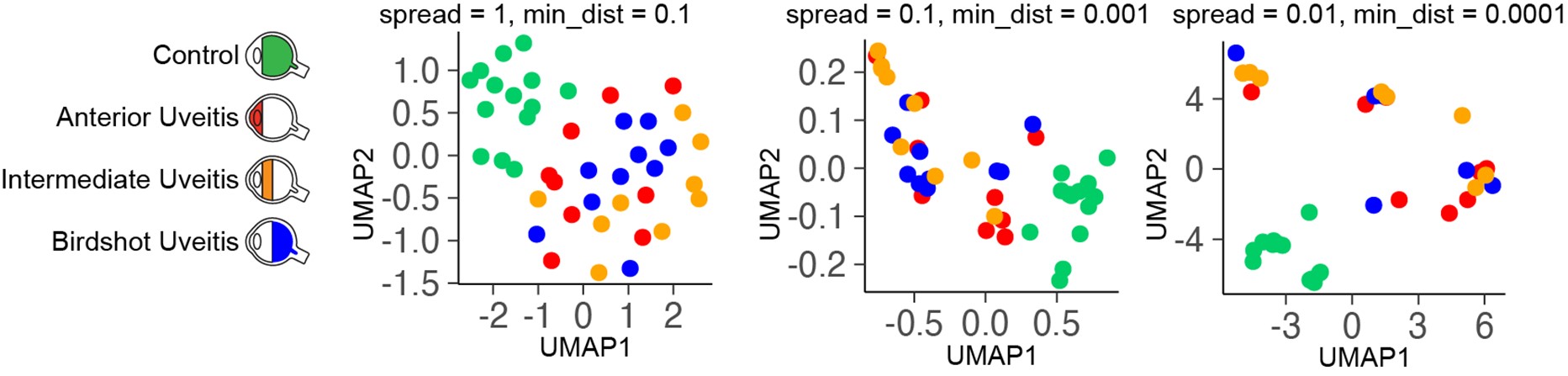
The use of principal component analysis for clustering may be underpowered; I would suggest the authors apply UMAP to determine if higher dimensional component analyses correlate with disease type.
Upon request of the reviewer, we have conducted UMAP (with different tuning of hyperparameters) on the DEGs (cohort I, see image below). We believe that UMAP analysis did not provide additional insights or correlates with disease type. We hope the reviewer agrees.
The false-discovery rate in large transcriptomic projects is challenging. While the authors are to be commended for employing a validation set, it would be useful to employ a Monte Carlo simulation in which groups are arbitrarily relabeled to determine the number of expected false discoveries within this data set (i.e. akin to Significance Analysis of Microarray techniques).
We determined the adjusted P values via the DESeq2 package (for false-discovery rate of 5% and Benjamini-Hochberg Procedure). The results are shown in Supplemental File 1K-1M and analysis in Figure 1A.
I do not fully understand the significance of the mouse CD11c-Runx3delta mice. It appears these data were derived from previous datasets or from bone marrow stromal line cultures. Did the authors attempt to generate autoimmune uveitis (i.e. EAU) in these animals? Without this the relevance for uveitis is unclear.
We did not attempt to induce experimental autoimmune uveitis in CD11c-Runx3delta mice. We used transcriptomic data from dendritic cells purified from this model to show that loss of RUNX3 induces a gene signature highly reminiscent of the gene module identified in non-infectious uveitis patients. Using enrichment analysis, we show that the transcriptome of patients is highly enriched for this signature which indicates that the decreased RUNX3 observed in patients underlies the upregulation of CD36, CX3CR1 and other surface genes. In other words, we used data from transgenic models to dissect which of the altered transcription factors were driving this gene module and we identified the RUNX3-NOTCH2 axis as an important contributor.
-

Evaluation Summary:
The authors provide evidence suggesting the gene expression profile of a specific subset of dendritic cells define features of specific forms of non-infectious uveitis. This work suggests specific pathways in these cells that may have mechanistic import in inflammatory eye disease. This manuscript is of interested to immunologists studying autoimmunity and ocular immunity. While the paper is largely descriptive, the data it presents should serve as a valuable resource for generating hypotheses about the pathogenesis of ocular autoimmune disease.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #1 agreed to share their name with the authors.)
-

Reviewer #1 (Public Review):
In the article "Whole transcriptome-sequencing and network analysis of CD1c+ human dendritic cells identifies cytokine-secreting subsets linked to type I IFN-negative autoimmunity to the eye," Hiddingh, Pandit, Verhagen, et al., analyze peripheral antigen presenting cells from patients with active uveitis and control patients, and find several differentially expressed transcription factors and surface markers. In addition, they find a subset of antigen presenting cells that is decreased in frequency in patients with uveitis that in previous publications was shown to be increased in the eye of patients with active uveitis. The greatest strength of this paper is the ability to obtain such a large number of samples from active uveitis patients that are not currently on systemic therapy. While the validation …
Reviewer #1 (Public Review):
In the article "Whole transcriptome-sequencing and network analysis of CD1c+ human dendritic cells identifies cytokine-secreting subsets linked to type I IFN-negative autoimmunity to the eye," Hiddingh, Pandit, Verhagen, et al., analyze peripheral antigen presenting cells from patients with active uveitis and control patients, and find several differentially expressed transcription factors and surface markers. In addition, they find a subset of antigen presenting cells that is decreased in frequency in patients with uveitis that in previous publications was shown to be increased in the eye of patients with active uveitis. The greatest strength of this paper is the ability to obtain such a large number of samples from active uveitis patients that are not currently on systemic therapy. While the validation experiments have methodologic flaws that decrease their usefulness, this study will still serve as a valuable resource in generating hypotheses about the pathogenesis of uveitis that can be tested in future projects.
Minor concerns:
Consistency between experiments: In Figure 1F only ~15% of the CD1c+ cells are CD14+, while in Figure 4 D shows that 100% of CD36 CX3CR1 DP cells are CD14 positive (and half the CD36+CX3CR1- cells), and the DP account for 30% of CD1c DCs.Since all CD36+CX3CR1+ cells are CD14+ (Figure 4D), how CX3CR1 ended up being differentially regulated in a similar way despite this population was excluded from 2nd bulk RNAseq data set should be commented on by the authors
Line 153: "...substantiates this gene set as a core transcriptional feature of human autoimmune uveitis." It would be difficult to argue that when only 137 of the 1236 DEGs from the first module are repeated in a validation data set that this is the core transcriptions set that defines the population in any uveitis. Further concerns include that the validation data set is not the same population, but rather a subset not containing CD14.
Line 220: Notch-dll experiments: with the experiments presented it is not possible to say that the changes are due to maintenance of CD1c+ DCs without further experiments outlining what NOTCH2 signaling changes throughout time. Is the population fully developed in the first 7 days of culture prior to adding NOTCH2 or ADAM10 inhibitors? Is there more apoptosis in this pathway? Less proliferation? It would be more accurate to say that there are fewer cDC2s after 14 days of culture without speculating the cause. In this experiment it is unclear why the gate of CD141/CD1c was chosen, as this appears to be in the middle of the population. In normal PBMCs CD141+ DCs would be CD1c negative; therefore why exclude the CD141hiCD1c+ and CD141loCD1c+ populations?
Line 256: The hypothesis that the loss of CD36+CX3CR1+ cells was due to migration to the eye doesn't make sense based on volume and number of cells. 0.1% of all PBMC is ~1x107 cells, and distributed throughout the eye would give about 1.3x106 cells/mL of eye volume. This would make the eye turbid which is not consistent with birdshot chorioretinopathy and would be rare in HLA-B27 anterior uveitis and intermediate uveitis.
Line 267: Would have liked to see the gating of CX3CR1/CD36 cells be more consistent (there are overlapping CX3CR1+ and CX3CR1- populations in 5A, but in Figure 4 quadrants were used to define the populations when evaluating the numbers in uveitis and healthy controls. The populations in Figure 5 are more separated by CD36.
Line 269, IN VITRO stimulation: The experimental paradigm is set up to find a difference between cells but does not to test any biologically relevant scenario. By sorting on a surface marker, then stimulating with the ligand for that receptor, the result better proves that CD36 is important in TLR2 signaling than does it give any information on how these dendritic cells might behave in uveitis.
-

Reviewer #2 (Public Review):
dendritic cells (DCs) are critical for initiating immune response and can initiate or accelerate autoimmune diseases. The authors are trying to better define the CD1c population in human autoimmune uveitis which was previously indicated to be involved in the disease. Thus, it is important to understand the role of dendritic cells in autoimmune uveitis.
the authors identified a population of CD1c in blood based on the combination of markers CD36 and CX3CR1 that is reduced in Uveitis.
the reviewer appreciate the difficulty in getting these patient samples. however, there are some technical issues with the analysis of the cells and lack of sufficient functional assays prevent the deep understanding of the biology of these cells.
if their reduction contributes to the diseases than a better understanding how to …
Reviewer #2 (Public Review):
dendritic cells (DCs) are critical for initiating immune response and can initiate or accelerate autoimmune diseases. The authors are trying to better define the CD1c population in human autoimmune uveitis which was previously indicated to be involved in the disease. Thus, it is important to understand the role of dendritic cells in autoimmune uveitis.
the authors identified a population of CD1c in blood based on the combination of markers CD36 and CX3CR1 that is reduced in Uveitis.
the reviewer appreciate the difficulty in getting these patient samples. however, there are some technical issues with the analysis of the cells and lack of sufficient functional assays prevent the deep understanding of the biology of these cells.
if their reduction contributes to the diseases than a better understanding how to boost it would be beneficial and impactful.
-

Reviewer #3 (Public Review):
First, a note on nomenclature. The authors use the term 'auto-immune' uveitis to encapsulate three different conditions -- HLA-B27 anterior uveitis, idiopathic intermediate uveitis, and birdshot choroidopathy. While I would agree with this terminology for the third set, there is substantial controversy as to whether HLA-B27 is truly autoimmune or autoinflammatory. Indeed, one major hypothesis is that this condition is driven by changes in gut microbiome. Intermediate uveitis is even more problematic; a substantial number of cases of this condition will turn out to be associated with demyelinating disease, which has recently been linked to Epstein Barr virus disease. To my knowledge in none of these diseases has a definitive autoantigen been identified nor passive transfer via transfusion shown; I would …
Reviewer #3 (Public Review):
First, a note on nomenclature. The authors use the term 'auto-immune' uveitis to encapsulate three different conditions -- HLA-B27 anterior uveitis, idiopathic intermediate uveitis, and birdshot choroidopathy. While I would agree with this terminology for the third set, there is substantial controversy as to whether HLA-B27 is truly autoimmune or autoinflammatory. Indeed, one major hypothesis is that this condition is driven by changes in gut microbiome. Intermediate uveitis is even more problematic; a substantial number of cases of this condition will turn out to be associated with demyelinating disease, which has recently been linked to Epstein Barr virus disease. To my knowledge in none of these diseases has a definitive autoantigen been identified nor passive transfer via transfusion shown; I would suggest the authors abandon this terminology and simply refer to the conditions as they are called.
Further, it would have been very desirable to compare the DC transcriptome for the other class of uveitic disease -- infectious -- for acute retinal necrosis or similar. As well it would have been very useful to compare profiles to other, related immune-mediated diseases such as ankylosing spondylitis.
Finally, it must be noted that looking for systemic signals in dendritic gene expression may be a bit of a needle in the haystack approach. Presumably, the function of the dendritic cells in uveitis is largely centered on those cells in the eye. It would have been highly desirable to examine the expression profile of intraocular DCs in at least a subset of patients who may have come to surgery (for instance, steroid implantation or vitrectomy).
It is also problematic that no effort has been made to assess the severity of uveitis. Flares of disease can range from extremely mild to debilitating. Similarly, intermediate uveitis and BSCR can range greatly in severity. Without normalizing for disease severity it is difficult to fully understand the range of transcriptional changes between cases.
The results are intriguing, but several additional analyses would further support these findings.
1. The use of principal component analysis for clustering may be underpowered; I would suggest the authors apply UMAP to determine if higher dimensional component analyses correlate with disease type.
2. The false-discovery rate in large transcriptomic projects is challenging. While the authors are to be commended for employing a validation set, it would be useful to employ a Monte Carlo simulation in which groups are arbitrarily relabeled to determine the number of expected false discoveries within this data set (i.e. akin to Significance Analysis of Microarray techniques).
3. I do not fully understand the significance of the mouse CD11c-Runx3delta mice. It appears these data were derived from previous datasets or from bone marrow stromal line cultures. Did the authors attempt to generate autoimmune uveitis (i.e. EAU) in these animals? Without this the relevance for uveitis is unclear.
-