Functional genomics reveals strain-specific genetic requirements conferring hypoxic growth in Mycobacterium intracellulare
Curation statements for this article:-
Curated by eLife
eLife Assessment
This study makes a valuable contribution by elucidating the genetic determinants of growth and fitness across multiple clinical strains of Mycobacterium intracellulare, an understudied non-tuberculous mycobacterium. Using transposon sequencing (Tn-seq), the authors identify a core set of 131 genes essential for bacterial adaptation to hypoxia, providing a convincing foundation for anti-mycobacterial drug discovery.
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Mycobacterium intracellulare is a major etiological agent of the recently expanding Mycobacterium avium–intracellulare complex pulmonary disease (MAC-PD). Therapeutic regimens that include a combination of macrolides and antituberculous drugs have been implemented with limited success. To identify novel targets for drug development that accommodate the genomic diversity of M. avium–intracellulare , we subjected eight clinical MAC-PD isolates and the type strain ATCC13950 to genome-wide profiling to comprehensively identify universally essential functions by transposon sequencing (TnSeq). Among these strains, we identified 131 shared essential or growth-defect-associated genes by TnSeq. Unlike the type strain, the clinical strains showed increased requirements for genes involved in gluconeogenesis and the type VII secretion system under standard growth conditions, the same genes required for hypoxic pellicle-type biofilm formation in ATCC13950. Consistent with the central role of hypoxia in the evolution of M. intracellulare , the clinical MAC-PD strains showed more rapid adaptation to hypoxic growth than the type strain. Importantly, the increased requirements of hypoxic fitness genes were confirmed in a mouse lung infection model. These findings confirm the concordant genetic requirements under hypoxic conditions in vitro and hypoxia-related conditions in vivo and highlight the importance of using clinical strains and host-relevant growth conditions to identify high-value targets for drug development.
Article activity feed
-

-

-

-

eLife Assessment
This study makes a valuable contribution by elucidating the genetic determinants of growth and fitness across multiple clinical strains of Mycobacterium intracellulare, an understudied non-tuberculous mycobacterium. Using transposon sequencing (Tn-seq), the authors identify a core set of 131 genes essential for bacterial adaptation to hypoxia, providing a convincing foundation for anti-mycobacterial drug discovery.
-

Reviewer #1 (Public review):
Summary:
In this descriptive study, Tateishi et al. report a Tn-seq based analysis of genetic requirements for growth and fitness in 8 clinical strains of Mycobacterium intracellulare Mi), and compare the findings with a type strain ATCC13950. The study finds a core set of 131 genes that are essential in all nine strains, and therefore are reasonably argued as potential drug targets. Multiple other genes required for fitness in clinical isolates have been found to be important for hypoxic growth in the type strain.
Strengths:
The study has generated a large volume of Tn-seq datasets of multiple clinical strains of Mi from multiple growth conditions, including from mouse lungs. The dataset can serve as an important resource for future studies on Mi, which despite being clinically significant, remains a …
Reviewer #1 (Public review):
Summary:
In this descriptive study, Tateishi et al. report a Tn-seq based analysis of genetic requirements for growth and fitness in 8 clinical strains of Mycobacterium intracellulare Mi), and compare the findings with a type strain ATCC13950. The study finds a core set of 131 genes that are essential in all nine strains, and therefore are reasonably argued as potential drug targets. Multiple other genes required for fitness in clinical isolates have been found to be important for hypoxic growth in the type strain.
Strengths:
The study has generated a large volume of Tn-seq datasets of multiple clinical strains of Mi from multiple growth conditions, including from mouse lungs. The dataset can serve as an important resource for future studies on Mi, which despite being clinically significant, remains a relatively understudied species of mycobacteria.
Weaknesses:
The primary claim of the study that the clinical strains are better adapted for hypoxic growth is yet to be comprehensively investigated. However, this reviewer thinks such an investigation would require a complex experimental design and perhaps form an independent study.
Comments on revisions:
The revised paper has satisfactorily addressed my previous concerns, and I have no further issues with this paper.
-

Author response:
The following is the authors’ response to the previous reviews
Reviewer #1 (Public review) :
Comments on revisions:
The revised manuscript has responded to the previous concerns of the reviewers, albeit modestly. The overemphasis on hypoxic adaptation of the clinical isolates persist as a key concern in the paper. The authors have compared the growth-curve of each of the clinical and ATCC strains under normal and hypoxic conditions (Fig. 8), but don't show how mutations in some of the genes identified in Tn-seq would impact the growth phenotype under hypoxia. They largely base their arguments on previously published results.
As I mentioned previously, the paper will be better without over-interpreting the TnSeq data in the context of hypoxia.
Thank you for the comment on the issue of not determining the impact of …
Author response:
The following is the authors’ response to the previous reviews
Reviewer #1 (Public review) :
Comments on revisions:
The revised manuscript has responded to the previous concerns of the reviewers, albeit modestly. The overemphasis on hypoxic adaptation of the clinical isolates persist as a key concern in the paper. The authors have compared the growth-curve of each of the clinical and ATCC strains under normal and hypoxic conditions (Fig. 8), but don't show how mutations in some of the genes identified in Tn-seq would impact the growth phenotype under hypoxia. They largely base their arguments on previously published results.
As I mentioned previously, the paper will be better without over-interpreting the TnSeq data in the context of hypoxia.
Thank you for the comment on the issue of not determining the impact of individual gene mutations identified in TnSeq on the growth phenotypes under hypoxia.
We agree that the lack of validation of TnSeq results is a limitation of this study. Without evidence of growth pattern of each gene-deletion mutant under hypoxia there might be a risk of over-interpretating the data, even though the data are carefully interpreted based on previous reports. We consider that it is necessary to confirm the phenomenon by using knockout mutants.
We have just recently succeeded in constructing the vector plasmids for making knockout mutants of M intracellulare (Tateishi. Microbiol Immunol. 2024). We will proceed to the validation experiment of TnSeq-hit genes by constructing knockout mutants. We already mentioned this point as a limitation of this study in the Discussion (pages 35-36 lines 630-640 in the revised manuscript).
Reference.
Tateishi, Y., Nishiyama, A., Ozeki, Y. & Matsumoto, S. Construction of knockout mutants in Mycobacterium intracellulare ATCC13950 strain using a thermosensitive plasmid containing negative selection marker rpsL+. Microbiol Immunol 68, 339-347 (2024).
Other points:
The y-axis legends of plots in Fig.8c are illegible.
Following the comment, we have corrected Figure 8c and checked the uploaded PDF
The statements in lines 376-389 are convoluted and need some explanation. If the clinical strains enter the log phase sooner than ATCC strain under hypoxia, then how come their growth rates (fig. 8c) are lower? Aren't they expected to grow faster?
Thank you for the comment on the interpretation of the difference in bacterial growth under hypoxia between MAC-PD strains and the ATCC type strain. The growth curve consists of the onset of logarithmic growth and its growth speed. In this study, we evaluated the former as timing of midpoint and the latter as growth rate at midpoint. Timing of midpoint and growth rate at midpoint are individual parameters. The early entry to log-phase does not mean the fast growth rate at midpoint.
Our results demonstrated that 5 (M.i.198, M.i.27, M003, M019 and M021) out of 8 clinical MAC-PD strains entered log-phase early and continued to grow logarithmically long time (slow growth). This data suggests the capacity for MAC-PD to continue replication long time under hypoxic conditions. By contrast, the ATCC type strain showed delayed onset of logarithmic growth caused by long-term lag phase. The duration of logarithmic growth was short even once after it started. The log phase soon transited to the stationary phase. This data suggests the lower capacity for the ATCC strain to continue replication under hypoxic conditions.
Following the comment, we have added the interpretation of the growth curve pattern as follows (page 22 lines 379-392 in the revised manuscript): “The growth rate at midpoint under hypoxic conditions was significantly lower in these 5 clinical MAC-PD strains than in ATCC13950. The early entry to log phase followed by long-term logarithmic growth (slow growth rate at midpoint) suggests the capacity for these 5 clinical MAC-PD strains to continue replication long time under hypoxic conditions. On the other hand, the rest 3 clinical MAC-PD strains (M018, M001 and MOTT64) did not show significant change in the growth rate between aerobic and hypoxic conditions, suggesting that there are different levels of capacity in maintaining long-term replication under hypoxia among clinical MAC-PD strains. In ATCC13950, the entry to log phase was significantly delayed under 5% oxygen compared to aerobic conditions, and the growth rate at midpoint was significantly increased under hypoxic conditions compared to aerobic conditions in ATCC13950. Such long-term lag phase followed by short-term log phase suggests lower capacity for ATCC13950 to continue replication under hypoxic conditions compared to clinical MAC-PD strains.”
Reviewer #4 (Public review):
Comments on revisions:
The revised version has satisfactorily addressed my initial comments in the discussion section.
The authors thank the Reviewer for understanding our reply.
Reviewer #5 (Public review):
Comments on revisions:
There is quite a lot of data and this could have been a really impactful study if the authors had channelized the Tn mutagenesis by focusing on one pathway or network. It looks scattered. However, from the previous version, the authors have made significant improvements to the manuscript and have provided comments that fairly address my questions.
The authors thank the Reviewer for understanding our reply. And the authors thank the Reviewer for the comments suggesting the future studies of TnSeq that focus on one pathway or network.
-

eLife Assessment
This study makes a valuable contribution by elucidating the genetic determinants of growth and fitness across multiple clinical strains of Mycobacterium intracellulare, an understudied non-tuberculous mycobacterium. Using transposon sequencing (Tn-seq), the authors identify a core set of 131 genes essential for bacterial adaptation to hypoxia, providing a convincing foundation for anti-mycobacterial drug discovery. Minor concerns remain regarding the presentation of Fig. 8C and the interpretation of data related to hypoxia.
-

Reviewer #1 (Public review):
Summary:
In this descriptive study, Tateishi et al. report a Tn-seq based analysis of genetic requirements for growth and fitness in 8 clinical strains of Mycobacterium intracellulare Mi), and compare the findings with a type strain ATCC13950. The study finds a core set of 131 genes that are essential in all nine strains, and therefore are reasonably argued as potential drug targets. Multiple other genes required for fitness in clinical isolates have been found to be important for hypoxic growth in the type strain.
Strengths:
The study has generated a large volume of Tn-seq datasets of multiple clinical strains of Mi from multiple growth conditions, including from mouse lungs. The dataset can serve as an important resource for future studies on Mi, which despite being clinically significant remains a …
Reviewer #1 (Public review):
Summary:
In this descriptive study, Tateishi et al. report a Tn-seq based analysis of genetic requirements for growth and fitness in 8 clinical strains of Mycobacterium intracellulare Mi), and compare the findings with a type strain ATCC13950. The study finds a core set of 131 genes that are essential in all nine strains, and therefore are reasonably argued as potential drug targets. Multiple other genes required for fitness in clinical isolates have been found to be important for hypoxic growth in the type strain.
Strengths:
The study has generated a large volume of Tn-seq datasets of multiple clinical strains of Mi from multiple growth conditions, including from mouse lungs. The dataset can serve as an important resource for future studies on Mi, which despite being clinically significant remains a relatively understudied species of mycobacteria.
Weaknesses:
The primary claim of the study that the clinical strains are better adapted for hypoxic growth is yet to be comprehensively investigated. However, this reviewer thinks such an investigation would require a complex experimental design and perhaps forms an independent study.
Comments on revisions:
The revised manuscript has responded to the previous concerns of the reviewers, albeit modestly. The overemphasis on hypoxic adaptation of the clinical isolates persist as a key concern in the paper. The authors have compared the growth-curve of each of the clinical and ATCC strains under normal and hypoxic conditions (Fig. 8), but don't show how mutations in some of the genes identified in Tn-seq would impact the growth phenotype under hypoxia. They largely base their arguments on previously published results.
As I mentioned previously, the paper will be better without over-interpreting the TnSeq data in the context of hypoxia.
Other points:
The y-axis legends of plots in Fig.8c are illegible.
The statements in lines 376-389 are convoluted and need some explanation. If the clinical strains enter the log phase sooner than ATCC strain under hypoxia, then how come their growth rates (fig. 8c) are lower? Aren't they are expected to grow faster?
-

Reviewer #4 (Public review):
Summary:
In this study Tateishi et al. used TnSeq to identify 131 shared essential or growth defect-associated genes in eight clinical MAC-PD isolates and the type strain ATCC13950 of Mycobacterium intracellulare which are proposed as potential drug targets. Genes involved in gluconeogenesis and the type VII secretion system which are required for hypoxic pellicle-type biofilm formation in ATCC13950 also showed increased requirement in clinical strains under standard growth conditions. These findings were further confirmed in a mouse lung infection model.
Strengths:
This study has conducted TnSeq experiments in reference and 8 different clinical isolates of M. intracellulare thus producing large number of datasets which itself is a rare accomplishment and will greatly benefit the research community.
Weaknesse…
Reviewer #4 (Public review):
Summary:
In this study Tateishi et al. used TnSeq to identify 131 shared essential or growth defect-associated genes in eight clinical MAC-PD isolates and the type strain ATCC13950 of Mycobacterium intracellulare which are proposed as potential drug targets. Genes involved in gluconeogenesis and the type VII secretion system which are required for hypoxic pellicle-type biofilm formation in ATCC13950 also showed increased requirement in clinical strains under standard growth conditions. These findings were further confirmed in a mouse lung infection model.
Strengths:
This study has conducted TnSeq experiments in reference and 8 different clinical isolates of M. intracellulare thus producing large number of datasets which itself is a rare accomplishment and will greatly benefit the research community.
Weaknesses:
(1) Comparative growth study of pure and mixed cultures of clinical and reference strains under hypoxia will be helpful in supporting the claim that clinical strains adapt better to such conditions. This should be mentioned as future directions in the discussion section along with testing the phenotype of individual knockout strains.
(2) Authors should provide the quantitative value of read counts for classifying a gene as "essential" or "non-essential" or "growth-defect" or "growth-advantage". Merely mentioning "no insertions in all or most of their TA sites" or "unusually low read counts" or "unusually high low read counts" is not clear.
(3) One of the major limitations of this study is the lack of validation of TnSeq results with individual gene knockouts. Authors should mention this in the discussion section.
Comments on revisions:
The revised version has satisfactorily addressed my initial comments in the discussion section.
-

Reviewer #5 (Public review):
Summary:
In the research article, "Functional genomics reveals strain-specific genetic requirements conferring hypoxic growth in Mycobacterium intracellulare" Tateshi et al focussed their research on pulmonary disease caused by Mycobacterium avium-intracellulare complex which has recently become a major health concern. The authors were interested in identifying the genetic requirements necessary for growth/survival within host and used hypoxia and biofilm conditions that partly replicate some of the stress conditions experienced by bacteria in vivo. An important finding of this analysis was the observation that genes involved in gluconeogenesis, type VII secretion system and cysteine desulphurase were crucial for the clinical isolates during standard culture while the same were necessary during hypoxia in …
Reviewer #5 (Public review):
Summary:
In the research article, "Functional genomics reveals strain-specific genetic requirements conferring hypoxic growth in Mycobacterium intracellulare" Tateshi et al focussed their research on pulmonary disease caused by Mycobacterium avium-intracellulare complex which has recently become a major health concern. The authors were interested in identifying the genetic requirements necessary for growth/survival within host and used hypoxia and biofilm conditions that partly replicate some of the stress conditions experienced by bacteria in vivo. An important finding of this analysis was the observation that genes involved in gluconeogenesis, type VII secretion system and cysteine desulphurase were crucial for the clinical isolates during standard culture while the same were necessary during hypoxia in the ATCC type strain.
Strength of the study:
Transposon mutagenesis has been a powerful genetic tool to identify essential genes/pathways necessary for bacteria under various in vitro stress conditions and for in vivo survival. The authors extended the TnSeq methodology not only to the ATCC strain but also to the recently clinical isolates to identify the differences between the two categories of bacterial strains. Using this approach they dissected the similarities and differences in the genetic requirement for bacterial survival between ATCC type strains and clinical isolates. They observed that the clinical strains performed much better in terms of growth during hypoxia than the type strain. These in vitro findings were further extended to mouse infection models and similar outcomes were observed in vivo further emphasising the relevance of hypoxic adaptation crucial for the clinical strains which could be explored as potential drug targets.
Weakness:
The authors have performed extensive TnSeq analysis but fail to present the data coherently. The data could have been well presented both in Figures and text. In my view this is one of the major weakness of the study.
Comments on revisions:
There is quite a lot of data and this could have been a really impactful study if the the authors had channelized the Tn mutagenesis by focussing on one pathway or network. It looks scattered. However, from the previous version, the authors have made significant improvements to the manuscript and have provided comments that fairly address my questions.
-

Author response:
The following is the authors’ response to the previous reviews.
Reviewer #1 (Public review):
Summary:
In this descriptive study, Tateishi et al. report a Tn-seq based analysis of genetic requirements for growth and fitness in 8 clinical strains of Mycobacterium intracellulare Mi), and compare the findings with a type strain ATCC13950. The study finds a core set of 131 genes that are essential in all nine strains, and therefore are reasonably argued as potential drug targets. Multiple other genes required for fitness in clinical isolates have been found to be important for hypoxic growth in the type strain.
Strengths:
The study has generated a large volume of Tn-seq datasets of multiple clinical strains of Mi from multiple growth conditions, including from mouse lungs. The dataset can serve as an important resource for …
Author response:
The following is the authors’ response to the previous reviews.
Reviewer #1 (Public review):
Summary:
In this descriptive study, Tateishi et al. report a Tn-seq based analysis of genetic requirements for growth and fitness in 8 clinical strains of Mycobacterium intracellulare Mi), and compare the findings with a type strain ATCC13950. The study finds a core set of 131 genes that are essential in all nine strains, and therefore are reasonably argued as potential drug targets. Multiple other genes required for fitness in clinical isolates have been found to be important for hypoxic growth in the type strain.
Strengths:
The study has generated a large volume of Tn-seq datasets of multiple clinical strains of Mi from multiple growth conditions, including from mouse lungs. The dataset can serve as an important resource for future studies on Mi, which despite being clinically significant remains a relatively understudied species of mycobacteria.
Thank you for the comment on the significance of our manuscript on the basic research of non-tuberculous mycobacteria.
Weaknesses:
The primary claim of the study that the clinical strains are better adapted for hypoxic growth is yet to be comprehensively investigated. However, this reviewer thinks such an investigation would require a complex experimental design and perhaps forms an independent study
Thank you for the comment on the issue of the claim of better adaptation for hypoxic growth in the clinical strains being not completely revealed. We agree the reviewer’s comment that comprehensive investigation of adaptation for hypoxic growth in the clinical strains should be a future project in terms of the complexity of an experimental design.
Reviewer #4 (Public review):
Summary:
In this study Tateishi et al. used TnSeq to identify 131 shared essential or growth defect-associated genes in eight clinical MAC-PD isolates and the type strain ATCC13950 of Mycobacterium intracellulare which are proposed as potential drug targets. Genes involved in gluconeogenesis and the type VII secretion system which are required for hypoxic pellicle-type biofilm formation in ATCC13950 also showed increased requirement in clinical strains under standard growth conditions. These findings were further confirmed in a mouse lung infection model.
Strengths:
This study has conducted TnSeq experiments in reference and 8 different clinical isolates of M. intracellulare thus producing large number of datasets which itself is a rare accomplishment and will greatly benefit the research community
Thank you for the comment on the significance of our manuscript on the basic research of non-tuberculous mycobacteria.
Weaknesses:
(1) A comparative growth study of pure and mixed cultures of clinical and reference strains under hypoxia will be helpful in supporting the claim that clinical strains adapt better to such conditions. This should be mentioned as future directions in the discussion section along with testing the phenotype of individual knockout strains.
Thank you for the comment on the idea of a comparative growth assay of pure and mixed cultures of clinical and reference strains under hypoxia. We appreciate the idea that showing the phenomenon of advantage of bacterial growth of the clinical strains under hypoxia in mixed culture with the ATCC strain would be important to strengthen the claim of better adaptation for hypoxic growth in the clinical strains. However, co-culture conditions introduce additional variables, including inter-strain competition or synergy, which can obscure the specific contributions of hypoxic adaptation in each strain. Therefore, we consider that our current approach using monoculture growth curves under defined oxygen conditions offers a clearer interpretation of strain-specific hypoxic responses.
Following the comment, we have added the mention of the mixed culture experiment and the growth assay using individual knockout strains as future directions (page 35 lines 614-632 in the revised manuscript).
“We have provided the data suggesting the preferential hypoxic adaptation in clinical strains compared to the ATCC type strain by the growth assay of individual strains. To strengthen our claim, several experiments are suggested including mixed culture experiments of clinical and reference strains under hypoxia. However, co-culture conditions introduce additional variables, including inter-strain competition or synergy, which can obscure the specific contributions of hypoxic adaptation in each strain. Therefore, we took the current approach using monoculture growth curves under defined oxygen conditions, which offers a clearer interpretation of strainspecific hypoxic responses. Furthermore, one of the limitations of this study is the lack of validation of TnSeq results with individual gene knockouts. Contrary to the case of Mtb, the technique of constructing knockout mutants of slow-growing NTM including M. intracellulare has not been established long time. We have just recently succeeded in constructing the vector plasmids for making knockout mutants of M intracellulare (Tateishi. Microbiol Immunol. 2024). Growth assay of individual knockout strains of genes showing increased genetic requirements such as pckA, glpX, csd, eccC5 and mycP5 in the clinical strains is suggested to provide the direct involvement of these genes on the preferential hypoxic adaptation in clinical strains. We have a future plan to construct knockout mutants of these genes to confirm the involvement of these genes on preferential hypoxic adaptation.”
Reference
Tateishi, Y., Nishiyama, A., Ozeki, Y. & Matsumoto, S. Construction of knockoutmutants in Mycobacterium intracellulare ATCC13950 strain using a thermosensitive plasmid containing negative selection marker rpsL+. Microbiol Immunol 68, 339-347 (2024).
(2) Authors should provide the quantitative value of read counts for classifying a gene as "essential" or "non-essential" or "growth-defect" or "growthadvantage". Merely mentioning "no insertions in all or most of their TA sites" or "unusually low read counts" or "unusually high low read counts" is not clear
Thank you for the comment on the issue of not providing the quantitative value of read counts for classifying the gene essentiality. In this study, we used an Hidden Markov Model (HMM) to predict gene essentiality. The HMM does not classify the 4 gene essentiality uniquely by the quantitative number of read counts but uses a probabilistic model to estimate the state at each TA based on the read counts and consistency with adjacent sites (Ioerger. Methods Mol Biol 2022).
The HMM uses consecutive data of read counts and calculates transition probability for predicting gene essentiality across the genome. The HMM allows for the clustering of insertion sites into distinct regions of essentiality across the entire genome in a statistically rigorous manner, while also allowing for the detection of growth-defect and growth-advantage regions. The HMM can smooth over individual outlier values (such as an isolated insertion in any otherwise empty region, or empty sites scattered among insertion in a non-essential region) and make a call for a region/gene that integrates information over multiple sites. The gene-level calls are made based on the majority call among the TA sites within each gene. The HMM automatically tunes its internal parameters (e.g. transition probabilities) to the characteristics of the input datasets (saturation and mean insertion counts) and can work over a broad range of saturation levels (as low as 20%) (DeJesus. BMC Bioinformatics 2013). Thus, HMM can represent the more nuanced ways the growth of an organism might be affected by the disruption of its genes (https://orca1.tamu.edu/essentiality/Tn-HMM/index.html)
Thus, the prediction of gene essentiality by the HMM does not rely on the quantitative threshold of Tn insertion reads independently at each TA site, but rather it is the most probable states for the whole sequence taken together (computed using Vitebri algorithm). Of the statistical methods, the HMM is a standard method for predicting gene essentiality in TnSeq (Ioerger TR. Methods Mol Biol. 2022) since a substantial number of TnSeq studies adopt this method for predicting gene essentiality (Akusobi. mBio 2025, DeJesus. mBio 2017, Dragset mSystems 2019, Mendum. BCG Genomics 2019). The HMM can be applied in many bioinformatics fields such as profiling functional protein families, identifying functional domains, sequence motif discoveries and gene prediction.
Taken together, we do not have the quantitative value of read counts for classifying gene essentiality by an HMM because the statistical methods for predicting gene essentiality do not uniquely use the quantitative value of read counts but use the transition of the read counts across the genome.
Reference
Ioerger TR. Analysis of Gene Essentiality from TnSeq Data Using Transit. Methods Mol Biol. 2022 ; 2377: 391–421. doi:10.1007/978-1-0716-1720-5_22.
DeJesus MA, Ioerger TR (2013) A Hidden Markov Model for identifying essential and 5 growth-defect regions in bacterial genomes from transposon insertion sequencing data. BMC Bioinformatics 14:303 [PubMed: 24103077]
Website by Ioerger: A Hidden Markov Model for identifying essential and growthdefect regions in bacterial genomes from transposon insertion sequencing data. https://orca1.tamu.edu/essentiality/Tn-HMM/index.html
Akusobi. C. et al. Transposon-sequencing across multiple Mycobacterium abscessus isolates reveals significant functional genomic diversity among strains. mBio 6, e0337624 (2025).
DeJesus, M.A. et al. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio 8, e02133-16 (2017).
Dragset, M.S., et al. Global assessment of Mycobacterium avium subsp. hominissuis genetic requirement for growth and virulence. mSystems 4, e00402-19 (2019). Mendum T.A., et al. Transposon libraries identify novel Mycobacterium bovis BCG genes involved in the dynamic interactions required for BCG to persist during in vivo passage in cattle. BMC Genomics 20, 431 (2019)
(3) One of the major limitations of this study is the lack of validation of TnSeq results with individual gene knockouts. Authors should mention this in the discussion section.
Thank you for the comment on the issue of the lack of validation of TnSeq results by using individual knockout mutants. We agree that the lack of validation of TnSeq results is one of the limitations of this study. We have just recently succeeded in constructing the vector plasmids for making knockout mutants of M intracellulare (Tateishi. Microbiol Immunol. 2024). We will proceed to the validation experiment of TnSeq-hit genes by constructing knockout mutants.
Following the comment, we have added the description in the Discussion (page 35 lines 622-632 in the revised manuscript) as follows: “Furthermore, one of the limitations of this study is the lack of validation of TnSeq results with individual gene knockouts. Contrary to the case of Mtb, the technique of constructing knockout mutants of slow-growing NTM including M. intracellulare has not been established long time. We have just recently succeeded in constructing the vector plasmids for making knockout mutants of M intracellulare (Tateishi. Microbiol Immunol 2024). Growth assay of individual knockout strains of genes showing increased genetic requirements such as pckA, glpX, csd, eccC5 and mycP5 in the clinical strains is suggested to provide the direct involvement of these genes on the 6 preferential hypoxic adaptation in clinical strains. We have a future plan to construct knockout mutants of these genes to confirm the involvement of these genes on preferential hypoxic adaptation.”
Reference
Tateishi, Y., Nishiyama, A., Ozeki, Y. & Matsumoto, S. Construction of knockout mutants in Mycobacterium intracellulare ATCC13950 strain using a thermosensitive plasmid containing negative selection marker rpsL + . Microbiol Immunol 68, 339-347 (2024).
Reviewer #5 (Public review):
Summary:
In the research article, "Functional genomics reveals strain-specific genetic requirements conferring hypoxic growth in Mycobacterium intracellulare" Tateshi et al focussed their research on pulmonary disease caused by Mycobacterium avium-intracellulare complex which has recently become a major health concern. The authors were interested in identifying the genetic requirements necessary for growth/survival within host and used hypoxia and biofilm conditions that partly replicate some of the stress conditions experienced by bacteria in vivo. An important finding of this analysis was the observation that genes involved in gluconeogenesis, type VII secretion system and cysteine desulphurase were crucial for the clinical isolates during standard culture while the same were necessary during hypoxia in the ATCC type strain.
Strength of the study:
Transposon mutagenesis has been a powerful genetic tool to identify essential genes/pathways necessary for bacteria under various in vitro stress conditions and for in vivo survival. The authors extended the TnSeq methodology not only to the ATCC strain but also to the recently clinical isolates to identify the differences between the two categories of bacterial strains. Using this approach they dissected the similarities and differences in the genetic requirement for bacterial survival between ATCC type strains and clinical isolates. They observed that the clinical strains performed much better in terms of growth during hypoxia than the type strain. These in vitro findings were further extended to mouse 7 infection models and similar outcomes were observed in vivo further emphasising the relevance of hypoxic adaptation crucial for the clinical strains which could be explored as potential drug targets.
Thank you for the comment on the significance of our manuscript on the basic research of non-tuberculous mycobacteria.
Weakness:
The authors have performed extensive TnSeq analysis but fail to present the data coherently. The data could have been well presented both in Figures and text. In my view this is one of the major weakness of the study.
Thank you for the comment on the issue of data presentation. Our point-by-point response to the Reviewer’s comments is shown below.
Reviewer #5 (Recommendations for the authors):
Major comments:
(1) The result section could have been better organized by splitting into multiple sections with each section focusing on a particular aspect.
Thank you for the comment on the organization of the section. We have split into multiple sections with each section focusing on a particular aspect as follows:
(1) Common essential and growth-defect-associated genes representing the genomic diversity of M. intracellulare strains (page 6 lines 102-103 in the revised manuscript)
(2) The sharing of strain-dependent and accessory essential and growth-defectassociated genes with genes required for hypoxic pellicle formation in the type strain ATCC13950 (page 8 lines 129-131 in the revised manuscript)
(3) Partial overlap of the genes showing increased genetic requirements in clinical MAC-PD strains with those required for hypoxic pellicle formation in the type strain ATCC13950 (page 9 lines 151-153 in the revised manuscript)
(4) Minor role of gene duplication on reduced genetic requirements in clinical MACPD strains (page 11 lines 184-185 in the revised manuscript)
(5) Identification of genes in the clinical MAC-PD strains required for mouse lung infection (page 12 lines 210-211 in the revised manuscript) 8
(6) Effects of knockdown of universal essential or growth-defect-associated genes in clinical MAC-PD strains (page 17 lines 305-306 in the revised manuscript)
(7) Differential effects of knockdown of accessory/strain-dependent essential or growth-defect-associated genes among clinical MAC-PD strains (page 19 lines 325- 326 in the revised manuscript)
(8) Preferential hypoxic adaptation of clinical MAC-PD strains evaluated with bacterial growth kinetics (page 21 lines 365-366 in the revised manuscript)
(9) The pattern of hypoxic adaptation not simply determined by genotypes (page 22 line 386 in the revised manuscript)
(2) The different strains that were used in the study, how they were isolated and some information on their genotypes could have been mentioned in brief in the main text and a table of different strains included as a supplementary table
Thank you for the comment on the information on the clinically isolated strains used in this study. All clinical strains were isolated from sputum of MAC-PD patients (Tateishi. BMC Microbiol. 2021, BMC Microbiol. 2023). Sputum samples were treated by the standard method for clinical isolation of mycobacteria with 0.5% (w/v) Nacetyl-L-cysteine and 2% (w/v) sodium hydroxide and plated on 7H10/OADC agar plates. Single colonies were picked up for use in experiments as isolated strains.
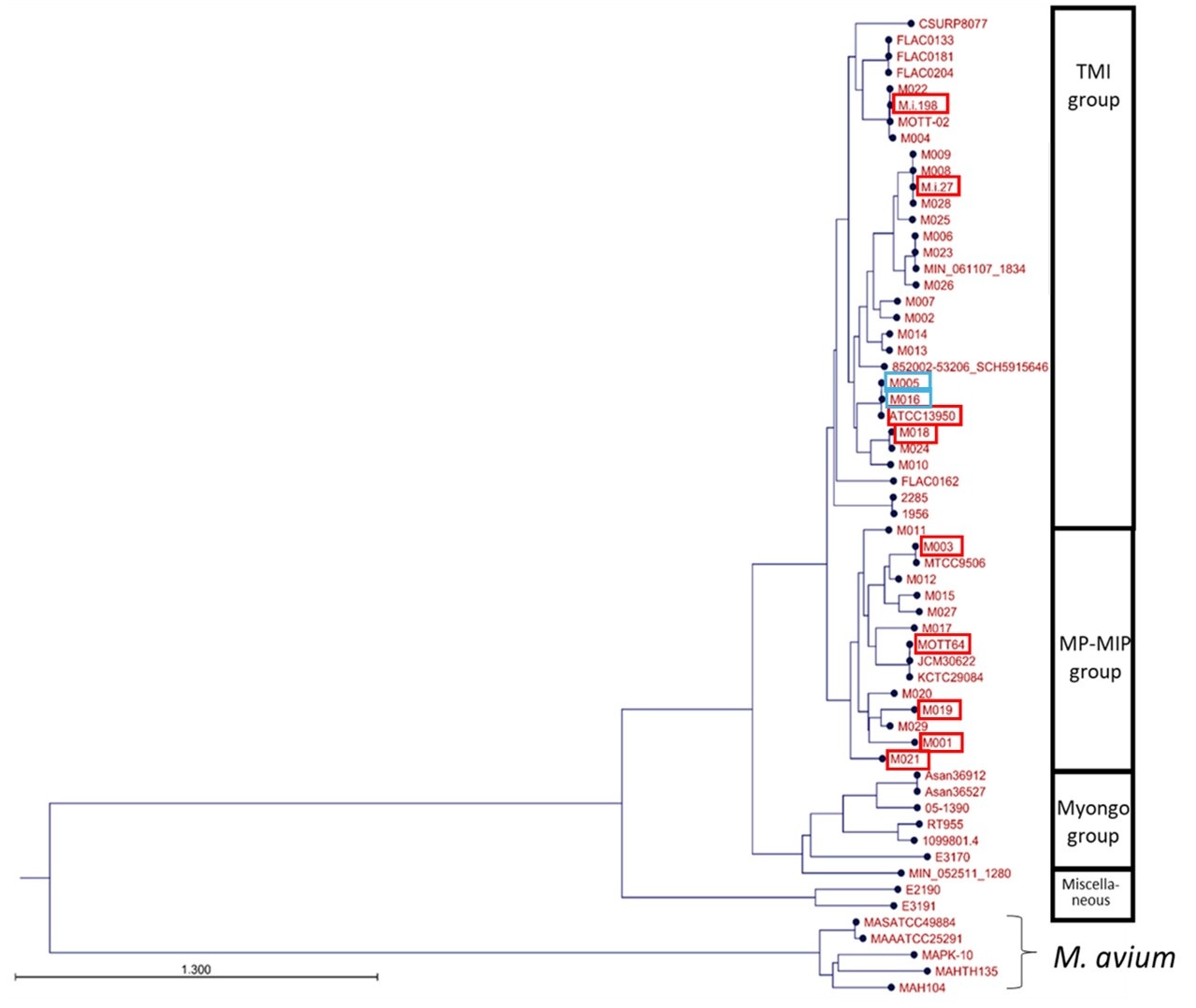
Following the comment, we have added the description on the information of the strains (page 37 lines 652-660 in the revised manuscript). “All eleven clinical strains from MAC-PD patients in Japan were isolated from sputum (Tateishi. BMC Microbiol 2021, BMC Microbiol 2023). Sputum samples were treated by the standard method for clinical isolation of mycobacteria with 0.5% (w/v) N-acetyl-L-cysteine and 2% (w/v) sodium hydroxide and plated on 7H10/OADC agar. Single colonies were picked up for use in experiments as isolated strains. Of these strains, ATCC13950, M.i.198, M.i.27, M018, M005 and M016 belong to the typical M. intracellulare (TMI) genotype and M001, M003, M019, M021 and MOTT64 belong to the M. paraintracellulare-M. indicus pranii (MP-MIP) genotype (Fig. 1, new Supplementary Table 1)”
Moreover, we have added the Supplementary Table showing the information on genotypes of each strain and the purpose of the use of study strains as new Supplementary Table 1
References
Tateishi, Y. et al. Comparative genomic analysis of Mycobacterium intracellulare: implications for clinical taxonomic classification in pulmonary Mycobacterium aviumintracellulare complex disease. BMC Microbiol 21, 103 (2021). Tateishi, Y. et al. Virulence of Mycobacterium intracellulare clinical strains in a mouse model of lung infection - role of neutrophilic inflammation in disease severity. BMC Microbiol 23, 94 (2023).
(3) As stated by the previous reviews, an explanation for the variation in the Tn insertion across different strains has not been provided and how they derive conclusions when the Tn frequency was not saturating.
Thank you for the comment on how to predict gene essentiality from our TnSeq data under the variation in the Tn insertion reads with suboptimal levels of saturation without reaching full saturation of Tn insertion.
As for the overcome of the Tn insertion variation, we normalized data by using Beta-Geometric correction (BGC), a non-linear normalization method. BGC normalizes the datasets to fit an “ideal” geometric distribution with a variable probability parameter ρ, and BGC improves resampling by reducing the skew. On TRANSIT software, we set the replicate option as Sum to combine read counts. And we normalized the datasets by Beta-Geometric correction (BGC) to reduce variabilities and performed resampling analysis by using normalized datasets to compare the genetic requirements between strains.
Following the comment, we have explained the variation in the Tn insertion across different strains in the manuscript (pages 39-40, lines 700-708 in the revised manuscript). “The number of Tn insertion in our datasets varied between 1.3 to 5.8 million among strains. To reduce the variation in the Tn insertion across strains, we adopt a non-linear normalization method, Beta-Geometric correction (BGC). BGC normalizes the datasets to fit an “ideal” geometric distribution with a variable probability parameter ρ, and BGC improves resampling by reducing the skew. On TRANSIT software, we set the replicate option as Sum to combine read counts. And we normalized the datasets by BGC and performed resampling analysis by using normalized datasets to compare the genetic requirements between strains.”
As for the issue of saturation levels of Tn insertion in our Tn mutant libraries, we made a description in the Discussion in the 1st version of the revised manuscript (pages 33-35 lines 592-613 in the 2nd version of the revised manuscript). The saturation of our Tn mutant libraries became 62-79% as follows: ATCC13950: 67.6%, M001: 72.9%, M003: 63.0%, M018: 62.4%, M019: 74.5%, M.i.27: 76.6%, M.i.198: 68.0%, MOTT64: 77.6%, M021: 79.9% by combining replicates. That is, we calculated gene essentiality from the Tn mutant libraries with 62-79% saturation in each strain. The levels of saturation of transposon libraries in our study are similar to the very recent TnSeq anlaysis by Akusobi where 52-80% saturation libraries (so-called “high-density” transposon libraries) are used for HMM and resampling analyses (Supplemental Methods Table 1[merged saturation] in Akusobi. mBio. 2025). The saturation of Tn insertion in individual replicates of our libraries is also comparable to that reported by DeJesus (Table S1 in mBio 2017). Thus, we consider that our TnSeq method of identifying essential genes and detecting the difference of genetic requirements between clinical MAC-PD strains and ATCC13950 is acceptable.
As for the identification of essential or growth-defect-associated genes by an HMM analysis, we do not consider that we made a serious mistake for the classification of essentiality by an HMM method in most of the structural genes that encode proteins. Because, as DeJesus shows, the number essential genes identified by TnSeq are comparable in large genes possessing more than 10 TA sites between 2 and 14 TnSeq datasets, most of which seem to be structural genes (Supplementary Fig 2 in mBio 2017). If the reviewer intends to regard our libraries far less saturated due to the smaller replicates (n = 2 or 3) than the previous DeJesus’ and Rifat’s reports using 10-14 replicates obtained to acquire so-called “high-density” transposon libraries (DeJesus. mBio 2017, Rifat. mBio 2021), there is a possibility that not all genes could be detected as essential due to the incomplete 11 covering of Tn insertion at nonpermissive TA sites, especially the small genes including small regulatory RNAs. Even if this were the case, it would not detract from the findings of our current study
As for the identification of genetic requirements by a resampling analysis, we consider that our data is acceptable because we compared the normalized data between strains whose saturation levels are similar to the previous report by Akusobi with “high-density” transposon libraries as mentioned above.
References
DeJesus, M.A., Ambadipudi, C., Baker, R., Sassetti, C. & Ioerger, T.R. TRANSIT--A software tool for Himar1 TnSeq analysis. PLoS Comput Biol 11, e1004401 (2015). Akusobi. C. et al. Transposon-sequencing across multiple Mycobacterium abscessus isolates reveals significant functional genomic diversity among strains. mBio 6, e0337624 (2025).
DeJesus, M.A. et al. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio 8, e02133-16 (2017).
Rifat, D., Chen L., Kreiswirth, B.N. & Nuermberger, E.L.. Genome-wide essentiality analysis of Mycobacterium abscessus by saturated transposon mutagenesis and deep sequencing. mBio 12, e0104921 (2021).
(4) ATCC strain is missing in the mouse experiment.
Thank you for the comment on the necessity of setting ATCC13950 as a control strain of mouse TnSeq experiment. To set ATCC13950 as a control strain in mouse infection experiments would be ideal. However, we have proved that ATCC13950 is eliminated within 4 weeks of infection in mice (Tateishi. BMC Microbiol 2023). To perform TnSeq, it is necessary to collect colonies at least the number of TA sites mathematically (Realistically, colonies with more than the number of TA sites are needed to produce biologically robust data.). That means, it is impossible to perform in vivo TnSeq study using ATCC13950 due to the inability to harvest sufficient number of colonies.
To make these things understood clearly, we have added the description of being unable to perform in vivo TnSeq in ATCC13950 in the result section (page 13 lines 221-222 in the revised manuscript).
“(It is impossible to perform TnSeq in lungs infected with ATCC13950 because ATCC13950 is eliminated within 4 weeks of infection) (Tateishi. BMC Microbiol 2023)”
Reference
Tateishi, Y. et al. Virulence of Mycobacterium intracellulare clinical strains in a mouse model of lung infection - role of neutrophilic inflammation in disease severity. BMC Microbiol 23, 94 (2023).
(5) The viability assays done in 96 well plate may not be appropriate given that mycobacterial cultures often clump without vigorous shaking. How did they control evaporation for 10 days and above?
Thank you for the comment on the issue of viability assay in terms of bacterial clumping. As described in the Methods (page 44 lines 778-781 in the revised manuscript), we have mixed the culture containing 250 μL by pipetting 40 times to loosen clumping every time before sampling 4 μL for inoculation on agar plates to count CFUs. By this method, we did not observe macroscopic clumping or pellicles like of Mtb or M. bovis BCG as seen in statistic culture.
We used inner wells for culture of bacteria in hypoxic growth assay. To control evaporation of the culture, we filled the distilled water in the outer wells and covered the plates with plastic lids. We cultured the plates with humidification at 37°C in the incubator.
(6) Fig. 7a many time points have only two data points and in few cases. The Y axis could have been kept same for better comparison for all strains and conditions.
Thank you for the comments on the data presentation of hypoxic growth assay in original Fig. 7a (new Fig 8a). The reason of many time points with only two data points is the close values of data in individual replicates. For example, the log10- transformed values of CFUs in ATCC13950 under aerobic culture are 4.716, 4.653, 4.698 at day 5, 4.949, 5.056, 4.954 at day 6, and 5.161, 5.190, 5.204 at day 8. We have added the numerical data of CFUs used for drawing growth curves as new Supplementary Table 19. Therefore, the data itself derives from three independent replicates.
Following the comment, we have revised the data presentation in new Fig 8a (original Fig. 7a) by keeping the same maximal value of Y axis across all graphs. In addition, we have revised the legend to designate clearly how we obtained the data of growth curves as follows (page 63 lines 1107-1108 in the revised manuscript): “Data on the growth curves are the means of three biological replicates from one experiment. Data from one experiment representative of three independent 13 experiments (N = 3) are shown.”
(7) The relevance of 7b is not well discussed and a suitable explanation for the difference in the profiles of M001 and MOTT64 between aerobic and hypoxia is not provided. Data representation should be improved for 7c with appropriate spacing.
Thank you for the comments on the relevance of original Fig. 7b (new Fig. 8b). In order to compare the pattern of logarithmic growth curves between strains quantitatively, we focused on time and slope at midpoint. The time at midpoint is the timing of entry to logarithmic growth phase. The earlier the strain enters logarithmic phase, the smaller the value of the time at midpoint becomes.
The two strains belonging to the MP-MIP subgroup, MOTT64 and M001 showed similar time at midpoint under aerobic conditions. However, the time at midpoint was significantly different between MOTT64 and M001 under hypoxia, the latter showing great delay of timing of entry to logarithmic phase. In contrast to the majority of the clinical strains that showed reduced growth rate at midpoint under hypoxia, neither strain showed such phenomenon under hypoxia. Although the implication in clinical situations has not been proven, strains without slow growth under hypoxia may have different (possibly strain-specific) mechanisms of hypoxic adaptation corresponding to the growth phenotypes under hypoxia.
Following the comment, we have added the explanation on the difference in the profiles of M001 and MOTT64 between aerobic and hypoxia in the Discussion (page 31 lines 552-557, page 32 lines 562-567 in the revised manuscript). “The two strains belonging to the MP-MIP subgroup, MOTT64 and M001 showed similar time at midpoint under aerobic conditions. However, the time at midpoint was significantly different between MOTT64 and M001 under hypoxia, the latter showing great delay of timing of entry to logarithmic phase. In contrast to the majority of the clinical strains that showed slow growth at midpoint under hypoxia, neither strain showed such phenomenon.”.
” Our inability to construct knockdown strains in M001 and MOTT64 prevented us from clarifying the factors that discriminate against the pattern of hypoxic adaptation. Although the implication in clinical situations has not been proven, strains without slow growth under hypoxia may have different (possibly strainspecific) mechanisms of hypoxic adaptation corresponding to the growth phenotypes under hypoxia.”
Following the comment, we have made the space between new Fig. 8b and 14 new Fig. 8c (original Fig. 7b and Fig. 7c).
(8) Fig. 8a, the antibiotic sensitivity at early and later time points do not seem to correlate. Any explanation?
Thank you for the comment on the uncorrelation of data of growth inhibition in knockdown strains of universal essential genes between early and later time points. The diminished effects of growth inhibition observed at Day 7 in knockdown strains may be due to the “escape” clones of knockdown strains under long-term culture by adding anhydrotetracycline (aTc) that induces sgRNA. As described in the Methods (pages 42-43 lines 754-758), we added aTc repeatedly every 48 h to maintain the induction of dCas9 and sgRNAs in experiments that extended beyond 48 h (Singh. Nucl Acid Res 2016). Such phenomenon has been reported by McNeil (Antimicrob Agent Chem. 2019) showing the increase in CFUs by day 9 with 100 ng/mL aTc with bacterial growth being detected between 2 and 3 weeks. These phenotypes of “escape” mutants is considered to be attributed to the promotor responsiveness to aTc.
Nevertheless, except for gyrB in M.i.27, the effect of growth inhibition at Day 7 in knockdown strains of universal essential genes was 10-1 or less of comparative growth rates of knockdown strains to vector control strains (y-axis of original Fig. 8). In this study, we judged the positive level of growth inhibition as 10-1 or less of comparative growth rates of knockdown strains to vector control strains (y-axis of new Fig. 7). Thus, we consider that the CRISPR-i data overall validated the essentiality of these genes.
References
Singh A.K., et al. Investigating essential gene function in Mycobacterium tuberculosis using an efficient CRISPR interference system, Nucl Acid Res 44, e143 (2016) McNeil M.B. &, Cook, G.M. Utilization of CRISPR interference to validate MmpL3 as a drug target in Mycobacterium tuberculosis. Antimicrob Agent Chem 63, e00629-19 (2019)
(9) Fig. 8b and c very data representation could have been improved. Some strains used in 7 are missing. The authors refer to technical challenge with respect to M001. Is it the same for others as well (MOTT64). The interpretation of data in result and discussion section is difficult to follow. Is the data subjected to statistical analysis?
Thank you for the comment on data presentation in original Fig. 8b (new Fig 7b). As 15 mentioned in the Discussion (page 18 lines 316-31 in the revised manuscript), the reason of missing M001 and MOTT64 in CRISPR-i experiment in original Fig. 7 (new Fig. 8) was we were unable to construct the knockdown strains in M001 and MOTT64. We consider these are the same technical challenges between M001 and MOTT64.
Following the comment, we have added the explanation of the technical challenge with respect to M001 and MOTT64 in the Discussion (page 32 lines 561- 566 in the revised manuscript). ”Our inability to construct knockdown strains in M001 and MOTT64 prevented us from clarifying the factors that discriminate against the pattern of hypoxic adaptation. Although the implication in clinical situations has not been proven, strains without slow growth under hypoxia may have different (possibly strain-specific) mechanisms of hypoxic adaptation corresponding to the growth phenotypes under hypoxia.”
As for the interpretation of growth suppression in knockdown experiments described in original Fig. 8 (new Fig. 7), We judged the positive level of growth inhibition as 10-1 or less of comparative growth rates of knockdown strains to vector control strains (y-axis of new Fig. 7). We interpreted the results based on whether the level of growth inhibition was positive or not (i.e. the comparative growth rates of knockdown strains to vector control strains became below 10-1 or not). Since our aim was to investigate whether knockdown of the target genes in each strain leads to growth inhibition, we did not perform statistical analysis between strains or target genes.
The major weakness of the study is the organization and data representation. It became very difficult to connect the role of gluconeogenesis, secretion system and others identified by authors to hypoxia, pellicle formation. The authors may consider rephrasing the results and discussion sections.
Thank you for the comments on the issue of organization and data presentation. Following the comment, we have revised the manuscript to indicate the relevance of the role of gluconeogenesis, secretion system and others defined by us more clearly (page 23 lines 404-408 in the revised manuscript).
“Because the profiles of genetic requirements reflect the adaptation to the environment in which bacteria habits, it is reasonable to assume that the increase of genetic requirements in hypoxia-related genes such as gluconeogenesis (pckA, glpX), type VII secretion system (mycP5, eccC5) and cysteine desulfurase (csd) play an important role on the growth under hypoxia-relevant conditions in vivo.”
Following the comments, we have exchanged the order of data presentation as follows: in vitro TnSeq (pages 6-12 lines 102-208 in the revised manuscript) , Mouse TnSeq (pages 12-17 lines 210-303 in the revised manuscript), Knockdown experiment (pages 17-21 lines 305-363 in the revised manuscript), Hypoxic growth assay (pages 21-23 lines 365-408 in the revised manuscript).
In association with the exchange of the order of data presentation, we have changed the order of the contents of the Discussion as follows: Preferential carbohydrate metabolism under hypoxia such as pckA and glpX (pages 24-26 lines 424-466 in the revised manuscript), Cysteine desulfurase gene (csd) (pages 26-27 lines 467-482 in the revised manuscript), Conditional essential genes in vivo such as type VII secretion system (pages 27-28 lines 483-497 in the revised manuscript), Knockdown experiment (pages 28-30 lines 498-536 in the revised manuscript), Hypoxic growth pattern (pages 30-32 lines 537-571 in the revised manuscript), Failure of assay using PckA inhibitors (pages 32-33 lines 572-578 in the revised manuscript), Transformation efficiencies (page 33 lines 579-591 in the revised manuscript), Saturation of Tn insertion (pages 33-35 lines 592-613 in the revised manuscript), Suggested future experiment plan (pages 35-36 lines 614-632 in the revised manuscript).
-

-

-

eLife Assessment
This investigation presents a valuable contribution by elucidating the genetic determinants of growth and fitness across multiple clinical strains of Mycobacterium intracellulare, an understudied non-tuberculous mycobacterium. Employing transposon sequencing (Tn-seq), the authors identify a core set of 131 genes essential for bacterial viability, offering a solid foundation for anti-mycobacterial drug discovery. However, there are minor but nonetheless significant concerns about data organization, which need to be addressed for greater scientific impact.
-

Reviewer #1 (Public review):
Summary:
In this descriptive study, Tateishi et al. report a Tn-seq based analysis of genetic requirements for growth and fitness in 8 clinical strains of Mycobacterium intracellulare Mi), and compare the findings with a type strain ATCC13950. The study finds a core set of 131 genes that are essential in all nine strains, and therefore are reasonably argued as potential drug targets. Multiple other genes required for fitness in clinical isolates have been found to be important for hypoxic growth in the type strain.
Strengths:
The study has generated a large volume of Tn-seq datasets of multiple clinical strains of Mi from multiple growth conditions, including from mouse lungs. The dataset can serve as an important resource for future studies on Mi, which despite being clinically significant remains a …
Reviewer #1 (Public review):
Summary:
In this descriptive study, Tateishi et al. report a Tn-seq based analysis of genetic requirements for growth and fitness in 8 clinical strains of Mycobacterium intracellulare Mi), and compare the findings with a type strain ATCC13950. The study finds a core set of 131 genes that are essential in all nine strains, and therefore are reasonably argued as potential drug targets. Multiple other genes required for fitness in clinical isolates have been found to be important for hypoxic growth in the type strain.
Strengths:
The study has generated a large volume of Tn-seq datasets of multiple clinical strains of Mi from multiple growth conditions, including from mouse lungs. The dataset can serve as an important resource for future studies on Mi, which despite being clinically significant remains a relatively understudied species of mycobacteria.
Weaknesses:
The primary claim of the study that the clinical strains are better adapted for hypoxic growth is yet to be comprehensively investigated. However, this reviewer thinks such an investigation would require a complex experimental design and perhaps forms an independent study.
-

Reviewer #4 (Public review):
Summary:
In this study Tateishi et al. used TnSeq to identify 131 shared essential or growth defect-associated genes in eight clinical MAC-PD isolates and the type strain ATCC13950 of Mycobacterium intracellulare which are proposed as potential drug targets. Genes involved in gluconeogenesis and the type VII secretion system which are required for hypoxic pellicle-type biofilm formation in ATCC13950 also showed increased requirement in clinical strains under standard growth conditions. These findings were further confirmed in a mouse lung infection model.
Strengths:
This study has conducted TnSeq experiments in reference and 8 different clinical isolates of M. intracellulare thus producing large number of datasets which itself is a rare accomplishment and will greatly benefit the research community.
Weaknesse…
Reviewer #4 (Public review):
Summary:
In this study Tateishi et al. used TnSeq to identify 131 shared essential or growth defect-associated genes in eight clinical MAC-PD isolates and the type strain ATCC13950 of Mycobacterium intracellulare which are proposed as potential drug targets. Genes involved in gluconeogenesis and the type VII secretion system which are required for hypoxic pellicle-type biofilm formation in ATCC13950 also showed increased requirement in clinical strains under standard growth conditions. These findings were further confirmed in a mouse lung infection model.
Strengths:
This study has conducted TnSeq experiments in reference and 8 different clinical isolates of M. intracellulare thus producing large number of datasets which itself is a rare accomplishment and will greatly benefit the research community.
Weaknesses:
(1) A comparative growth study of pure and mixed cultures of clinical and reference strains under hypoxia will be helpful in supporting the claim that clinical strains adapt better to such conditions. This should be mentioned as future directions in the discussion section along with testing the phenotype of individual knockout strains.
(2) Authors should provide the quantitative value of read counts for classifying a gene as "essential" or "non-essential" or "growth-defect" or "growth-advantage". Merely mentioning "no insertions in all or most of their TA sites" or "unusually low read counts" or "unusually high low read counts" is not clear.
(3) One of the major limitations of this study is the lack of validation of TnSeq results with individual gene knockouts. Authors should mention this in the discussion section. -

Reviewer #5 (Public review):
Summary:
In the research article, "Functional genomics reveals strain-specific genetic requirements conferring hypoxic growth in Mycobacterium intracellulare" Tateshi et al focussed their research on pulmonary disease caused by Mycobacterium avium-intracellulare complex which has recently become a major health concern. The authors were interested in identifying the genetic requirements necessary for growth/survival within host and used hypoxia and biofilm conditions that partly replicate some of the stress conditions experienced by bacteria in vivo. An important finding of this analysis was the observation that genes involved in gluconeogenesis, type VII secretion system and cysteine desulphurase were crucial for the clinical isolates during standard culture while the same were necessary during hypoxia in …
Reviewer #5 (Public review):
Summary:
In the research article, "Functional genomics reveals strain-specific genetic requirements conferring hypoxic growth in Mycobacterium intracellulare" Tateshi et al focussed their research on pulmonary disease caused by Mycobacterium avium-intracellulare complex which has recently become a major health concern. The authors were interested in identifying the genetic requirements necessary for growth/survival within host and used hypoxia and biofilm conditions that partly replicate some of the stress conditions experienced by bacteria in vivo. An important finding of this analysis was the observation that genes involved in gluconeogenesis, type VII secretion system and cysteine desulphurase were crucial for the clinical isolates during standard culture while the same were necessary during hypoxia in the ATCC type strain.
Strength of the study:
Transposon mutagenesis has been a powerful genetic tool to identify essential genes/pathways necessary for bacteria under various in vitro stress conditions and for in vivo survival. The authors extended the TnSeq methodology not only to the ATCC strain but also to the recently clinical isolates to identify the differences between the two categories of bacterial strains. Using this approach they dissected the similarities and differences in the genetic requirement for bacterial survival between ATCC type strains and clinical isolates. They observed that the clinical strains performed much better in terms of growth during hypoxia than the type strain. These in vitro findings were further extended to mouse infection models and similar outcomes were observed in vivo further emphasising the relevance of hypoxic adaptation crucial for the clinical strains which could be explored as potential drug targets.
Weakness:
The authors have performed extensive TnSeq analysis but fail to present the data coherently. The data could have been well presented both in Figures and text. In my view this is one of the major weakness of the study.
-

Author response:
The following is the authors’ response to the original reviews
Public Reviews:
Reviewer #1 (Public Review):
Summary:
Tateishi et al. report a Tn-seq-based analysis of genetic requirements for growth and fitness in 8 clinical strains of Mycobacterium intracellulare Mi), and compare the findings with a type strain ATCC13950. The study finds a core set of 131 genes that are essential in all nine strains, and therefore are reasonably argued as potential drug targets. Multiple other genes required for fitness in clinical isolates have been found to be important for hypoxic growth in the type strain.
Strengths:
The study has generated a large volume of Tn-seq datasets of multiple clinical strains of Mi from multiple growth conditions, including from mouse lungs. The dataset can serve as an important resource for future …
Author response:
The following is the authors’ response to the original reviews
Public Reviews:
Reviewer #1 (Public Review):
Summary:
Tateishi et al. report a Tn-seq-based analysis of genetic requirements for growth and fitness in 8 clinical strains of Mycobacterium intracellulare Mi), and compare the findings with a type strain ATCC13950. The study finds a core set of 131 genes that are essential in all nine strains, and therefore are reasonably argued as potential drug targets. Multiple other genes required for fitness in clinical isolates have been found to be important for hypoxic growth in the type strain.
Strengths:
The study has generated a large volume of Tn-seq datasets of multiple clinical strains of Mi from multiple growth conditions, including from mouse lungs. The dataset can serve as an important resource for future studies on Mi, which despite being clinically significant remains a relatively understudied species of mycobacteria.
Thank you for reviewing our manuscript and finding the significance of our data.
Weaknesses:
The paper lacks clarity in data presentation and organization. For example, some of the key data on cfu counts of clinical Mi strains in a mouse model can be presented along with the Tn-seq dataset in Figure 6, the visualization of which can be improved with volcano plots. etc. Improvement in data visualization is perhaps necessary throughout the paper.
Thank you for the comment on the data presentation of in vivo studies. We previously revealed the time-course data on CFUs, animal survival, and tissue pathology from the pure strains (Tateishi Y. BMC Microbiol. 2023; new Ref #22) . Based on these data, we assumed that we would be able to harvest sufficient number of colonies from mice infected with M.i.27 or M.i.198, and we performed in vivo TnSeq studies using these two strains. We have referred to our previous publication (new Ref #22) on the virulence of MAC-PD strains used in this study for mice in the revised manuscript (page12, line 212).
The data of CFU counts were shown in new Supplementary Fig. 3b. In the manuscript text, we explained as follows (page 12, lines 212-216): “The time course of the changes in the bacterial burden showed a pattern similar to those of the wild-type strains M.i.198 and M.i.27, respectively, except that it was not possible to harvest sufficient colonies (as few as 104/mouse) in the few mice infected with the M.i.27 Tn mutant strain in week 8 and week 16 (page 12, lines 212-216; new Supplementary Fig, 3b, new Supplementary Table 8)”.
Regarding the suggestion to include volcano plots, we appreciate the proposal but chose not to adopt this format, as the main aim of this study was to identify genes commonly required for in vitro and in vivo fitness across multiple M. intracellulare strains, rather than to highlight differential genetic requirements within a single strain. Volcano plots are useful for visualizing differential values and significance for a single dataset but are less suited for cross-strain comparisons of shared gene sets. Our approach is aligned with the methodology used by Cary et al. (PLoS Pathog. 2018; new Ref#8), who similarly focused on identifying conserved genetic requirements across M. tuberculosis genotypes without employing volcano plots.
[References]
Tateishi, Y. et al. Virulence of Mycobacterium intracellulare clinical strains in a mouse model of lung infection - role of neutrophilic inflammation in disease severity. BMC Microbiol 23, 94 (2023).
Carey, A.F. et al. TnSeq of Mycobacterium tuberculosis clinical isolates reveals strain-specific antibiotic liabilities. PLoS Pathog 14, e1006939 (2018).
The primary claim of the study that the clinical strains are better adapted for hypoxic growth is not well-supported by the data presented in Figure 7.
Thank you for the comments on the difference of adaptation for hypoxic growth between ATCC13950 and clinical MAC-PD strains. To clarify, growth rates shown in Figure 7 were calculated at the inflection point (midpoint) of the growth curves, which were modeled using a four-parameter logistic (4P logistic) model. As described in the Discussion, we found the pattern of hypoxic adaptation characteristics of the clinical MAC-PD strains from the growth curve forms. Taking into consideration the impact of growing bacteria on the disease progression of MAC-PD, the slow growth with early entry to log phase implicates the continuous impact on the infected hosts during logarithmic bacterial growth, which may be involved in the persistent and steadily progressive illness of MAC-PD for years in humans.
Unlike time-lapse imaging assay, the completely seamless sampling of culture for CFU assay is impossible. Nevertheless, we collected sufficient timepoints to allow reliable curve fitting with the 4P logistic model and thus consider our growth data to represent a valid approximation of continuous growth dynamics.
Regarding the suggestion of mixed culture experiments, we agree that such studies could be informative. However, co-culture conditions introduce additional variables, including inter-strain competition or synergy, which can obscure the specific contributions of hypoxic adaptation in each strain. Therefore, we believe that the current approach using monoculture growth curves under defined oxygen conditions offers a clearer interpretation of strain-specific hypoxic responses.
The title of the paper is misleading as the study doesn't provide any mechanistic aspect of hypoxic adaptation in Mi.
Thank you for the comment on the article title. We admit that this paper does not directly reveal the mechanism of hypoxic adaptation in M. intracellulare strains but provides the data on the different pattern of hypoxic adaptation between M. intracellulare strains in relation to the difference of genetic requirements. Therefore, we revised the title as ”Functional genomics reveals strain-specific genetic requirements conferring hypoxic growth in Mycobacterium intracellulare”
Reviewer #2 (Public Review):
Summary:
In the study titled "Functional genomics reveals the mechanism of hypoxic adaptation in nontuberculous mycobacteria" by Tateishi et al., the authors have used TnSeq to identify the common essential and growth-defect-associated genes that represent the genomic diversity of clinical M. intracellulare strains in comparison to the reference type strain. By estimating the frequency of Tn insertion, the authors speculate that genes involved in gluconeogenesis, the type VII secretion system, and cysteine desulfurase are relatively critical in the clinical MAC-PD strains than in the type strain, both for the extracellular survival and in a mouse lung infection model.
Based on their analysis, the authors proposed to identify the mechanism of hypoxic adaptation in nontuberculous mycobacteria (NTM) which offer promising drug targets in the strains causing clinical Mycobacterium avium-intracellulare complex pulmonary disease (MAC-PD).
Strengths:
A major strength of the manuscript is the performance of the exhaustive set of TnSeq experiments with multiple strains of M. intracellulare during in vitro growth and animal infection.
Thank you for reviewing our manuscript and acknowledging the performance of producing datasets in this study.
Weaknesses:
(1) The study suffers from the authors' preconceived bias toward a small subset of genes involved in hypoxic pellicle formation in ATCC13950.
Thank you for the comment regarding a potential bias toward a small subset of genes involved in hypoxic pellicle formation in ATCC13950. The rationale for the importance of hypoxic pellicle genes in clinical MAC-PD strains is that the profiles of genetic requirements in each bacterial strain reflect the adaptation to the environment in which each strain lives. When the strains are placed in a special environment, they can adapt to the situation by altering the profiles of genetic requirements, resulting in the remodeling of metabolic pathways.
In this study, we found that several of these pellicle-associated genes also showed increased genetic requirement in the clinical MAC-PD strains, suggesting a possible overlap in hypoxic adaptation mechanisms. We did not insist that clinical MAC-PD strains showed an increase of genetic requirements in all genes of hypoxic pellicle formation. Except for the gene sets involved in hypoxic pellicle formation in ATCC13950, almost no global information has been revealed on the pathogenesis of nontuberculous mycobacterial disease, which differs from the case of tuberculosis. Along with this finding, we investigated the effect of gene silencing on bacterial growth and preferential hypoxic adaptation observed by growth kinetics in clinical MAC-PD strains compared to ATCC13950. At first glance, to focus on the gene sets of hypoxic pellicle formation seems to be “biased”, but we proceeded this research step by step based on our achievements. We consider these data provide valuable information on the pathogenesis of MAC-PD by clinical MAC-PD strains.
We have added the description of the rationale for the importance of hypoxic pellicle genes in clinical MAC-PD strains in the revised manuscript (page 9, lines 148-155).
(2) An important set of data with the ATCC13950 reference strain is missing in the mouse infection study. In the absence of this, it is difficult to establish whether the identified genes are critical for infection/intracellular proliferation, specifically in the clinical isolates that are relatively more adapted for hypoxia.
Thank you for the comment on the necessity of setting ATCC13950 as a control strain of mouse TnSeq experiment. To set ATCC13950 as a control strain in mouse infection experiments would be ideal. However, we proved that ATCC13950 is eliminated within 4 weeks of infection (Tateishi Y. BMC Microbiol. 2023; new Ref#22). That means, it is impossible to perform in vivo TnSeq study due to the inability to harvest sufficient number of colonies.
[Reference]
Tateishi, Y. et al. Virulence of Mycobacterium intracellulare clinical strains in a mouse model of lung infection - role of neutrophilic inflammation in disease severity. BMC Microbiol 23, 94 (2023).
(3) Statistical enrichment analysis of gene sets by GSEA wrongly involves genes required for hypoxic pellicle formation in ATCC13950 together with the gene sets found essential in the clinical MAC-PD strains, to claim that a significant % of genes belong to hypoxia-adaptation pathways. It could be factually incorrect because a majority of these might overlap with those found critical for the in vitro survival of MAC-PD strains (and may not be related to hypoxia).
Thank you for the suggestion on the re-analysis of gene enrichment analysis of genes required for M.i.27 and M.i.198 in vivo infection, individually with genes involved in hypoxic pellicle formation in ATCC13950 and with those showing increased genetic requirements in clinical MAC-PD strains compared to ATCC13950.
About 50% (92 and 94 out of 181 genes through Day 1 to Week 16 and Week4 to Week16 of infection) and 40% (70 and 79 genes out of 179 through Day 1 to Week 16 and Week 4 to Week 16 of infection) of genes required for hypoxic pellicle formation in ATCC13950 were listed as enriched in genes required for mouse lung infection in M.i.27 and M.i.198, respectively. In addition, about 42% (54 and 56 out of 128 genes through Day 1 to Week 16 and thorough Week 4 to Week 16 of infection) and 40% (79 and 68 out of 179 genes through Day 1 to Week 16 and through Week 4 to Week 16 of infection) of genes showing increased requirements in clinical MAC-PD strains compared to ATCC13950 were listed as enriched in genes required for mouse lung infection in M.i.27 and M.i.198, respectively.
These data indicate that about 40-50% genes required for in vitro hypoxic pellicle formation are shared with the genes required for in vivo bacterial growth, and that about 40% strain-dependent/accessory essential genes are shared with the genes required for in vivo bacterial growth. Thus, the genes required for the growth of M.i.27 and M.i.198 in mouse lungs are enriched individually with those involved in hypoxic pellicle formation in ATCC13950, and with the gene sets found critical for in vitro growth. We have added the description of the reanalyzed data of GSEA in the manuscript (pages 16-17, lines 287-290). And the details of reanalyzed data of GSEA have been shown in Supplementary Fig. 5 and 6 as well as Supplementary Tables 15 and 16.
(4) Validation of mouse infection experiments with individual mutants is missing.
Thank you for the suggestion on the validation of the TnSeq hit genes on the in vivo survival. We acknowledge the importance of validating the TnSeq-hit genes by constructing knockout mutants. We have recently succeeded in constructing the vectors for making knockout strains of M. intracellulare (Tateishi. Microbiol Immunol. 2024). We will proceed to the infection experiment of knockout mutants by using our system for constructing them.
[Reference]
Tateishi Y. et al. Construction of knockout mutants in Mycobacterium intracellulare ATCC13950 strain using a thermosensitive plasmid containing negative selection marker rpsL. Microbiol Immunol 68, 339-347 (2024).
(5) Phenotypes with TnSeq and CRISPRi-based KD exhibit poor correlation with misleading justifications by the authors.
Thank you for the comment on the issue of inconsistent results between TnSeq and CRISPR-i based knockdown. We acknowledge that some inconsistencies were observed, particularly among strain-dependent/accessory essential or growth-defect-associated genes. By contrast, we found consistent data between TnSeq and CRISPR-i based knockdown results among universal essential genes such as glcB, inhA, gyrB and embB. Although the mechanism has not been fully proven yet, we consider that such inconsistent phenotypes with TnSeq and CRISPR- based knockdown may be related to the recently revealed bypass mechanism of gene essentiality which is characteristically observed in strain-specific/accessory essential genes (Rosconi F. Nat Micorbiol. 2022; new Ref#14). They suggested this bypass mechanism of gene essentiality in strain-dependent/accessory essential or growth-defect-associated genes from the ‘forced-evolution experiments’ of 36 clinical Streptococcus pneumoniae strains. For example, knockout mutants are successfully recovered from transformation experiments targeting strain-specific/accessory essential genes in TnSeq such as cytidine monophosphate kinase cmk, formate tetrahydrofolate ligase fhs and farnesyl-diphosphate synthase fpp. The bypassing of gene essentiality can be suggested by observing suppressor mutations and synthetic lethality in knockout strains. By contrast, universal essential genes fulfill the following three categories: i) high levels of conservation within and often across species, iii) limited genetic diversity, and iii) high and stable expression levels. Consequently, the universal essential genes are rigid, largely immutable key components to an organism’s survival. In the universal essential genes, the knockout recovery fails as shown by no colonies or only appearance of merodiploids. Taking into consideration such bypass mechanism of gene essentiality in strain-dependent/accessory essential genes, the lower effect of gene silencing of strain-dependent/accessory essential genes on bacterial growth may reflect pathway rewiring that helps the bacterial growth under suppression of the target gene expression.
We have added the description of the possible reason for inconsistency between TnSeq and CRISPR-i results in the Result and Discussion in the revised manuscript (page 21, lines 367-376; pages 28-29, lines 489-519).
[Reference]
Rosconi, F. et al. A bacterial pan-genome makes gene essentiality strain-dependent and evolvable. Nat Microbiol 7, 1580–1592 (2022).
In summary, this study is unable to provide mechanistic insights into why and how different MAC-PD mutant strains exhibit differential survival (in vitro and in animals) and adaptation to hypoxia. It remains to understand why the clinical strains show better adaptation to hypoxia and what is the impact of other stresses on their growth rates.
Thank you for the comments on the issue of being unable to prove the mechanism of MAC-PD pathogenesis and adaptation to hypoxia. We admit that the original manuscript did not provide the apparent reason and mechanism of MAC-PD pathogenesis and adaptation to hypoxia. Following the comment, we have modified the manuscript tile as “Functional genomics reveals strain-specific genetic requirements conferring hypoxic growth in Mycobacterium intracellulare”.
However, we revealed the diversity of genetic requirements among the genus M. intracellulare including the type strain ATCC13950 and clinical MAC-PD strains. We revealed the characteristics of genetic requirements in clinical MAC-PD strains as increased genetic requirements of gluconeogenesis, type VII secretion system and cysteine desulfurase, the former two of which are also required in hypoxic pellicle formation in ATCC13950. Along with this, we demonstrated the difference of growth behavior under hypoxia between clinical MAC-PD strains and ATCC13950. Overall, we consider that we could provide the basic information suggesting the involvement of difference of genetic requirements among strains in the pathogenesis of MAC-PD.
Reviewer #3 (Public Review):
Summary:
The study by Tateishi et al. utilized TnSeq in nine genetically diverse M. intracellulare strains, identifying 131 common essential and growth-defect-associated genes across those strains, which could serve as potential drug targets. The authors also provided an overview of the differences in gene essentiality required for hypoxic growth between the reference strain and the clinical strains. Furthermore, they validated the universal and accessory/strain-dependent essential genes by knocking down their expression using CRISPRi technique. Overall, this study offers a comprehensive assessment of gene requirements in different clinical strains of M. intracellular.
Thank you for reviewing our manuscript and finding the significance of our data.
(1) The rationale for using ATCC13950 versus clinical strains needs to be clarified. The reference strain ATCC13950 was obtained from the abdominal lymph node of a patient around 10 years ago and is therefore considered a clinical strain that has undergone passages in vitro. How many mutations have accumulated during these in vitro passages? Are these mutations significant enough to cause the behavior of ATCC13950 to differ from other recently sampled clinical strains? From the phylogenetic tree, ATCC13950 is located between M018 and M.i.27. Did the authors observe a similarity in gene essentiality between ATCC13950 and its neighbor strains? What is the key feature that separates ATCC13950 from these clinical strains? The authors should provide a strong rationale for how to interpret the results of this comparison in a clinical or biological context.
Thank you for the comments on the rationale for using ATCC13950 versus clinical strains and the key feature that separates ATCC13950 from clinical MAC-PD strains.
ATCC13950 was isolated in 1949, (not around 10 years ago) from 34-month-old female abdominal lymph node (Cuttino. Am J Pathol 1949; new Ref#11). Of note, the clinical background of the patient infected with ATCC13950 is quite different from the patients with MAC-pulmonary disease (MAC-PD), the incidence rate of which has been increasing worldwide without predisposing immunological disorders. ATCC13950 has been regarded as a type strain of genus M. intracellulare historically. And ATCC13950 is the first M. intracellulare strain to be sequenced in 2012 (Kim. J Bacteriol 2012; new Ref#59).
The rationale for using ATCC13950 versus clinical MAC-PD strains is to find the answer to the question whether the essential genes and genetic requirements are similar or different between clinical MAC-PD strains and historical type strain ATCC13950. So far, there are two reports on TnSeq that compare genetic requirements between clinical mycobacterial strains and the type strains, one of which is M. tuberculosis (Carey AF. PLoS Pathogens. 2018; new Ref#8) and the other is M. abscessus (Akusobi C. mBio. 2025; new Ref#9, published after submission of our manuscript). They reported the difference and diversity of genetic requirements between clinical strain and type strains such as M. tuberculosis H37Rv and M. abscessus ATCC19977. We have added the mention of these previous reports to explain the rationale for setting the type strain ATCC13950 as a referential control strain. (page 5, lines 83-87)
The genetic and functional analysis of clinical MAC-PD strains has not been conducted for a long time. In 2021, we have revealed the genomic diversity between clinical MAC-PD and ATCC13950 by comparative genomic analysis (Tateishi BMC Microbiol. 2021; new Ref#5). Except for our TnSeq study of ATCC13950 (Tateishi Sci Rep 2020; new Ref#10), no functional analysis has been conducted in clinical M. intracellulare strains. On our research stream of clinical MAC-PD strains, we expected that we could reveal the functional genomic characteristics of clinical MAC-PD strains by setting ATCC13950 as a referential control strain for analyzing TnSeq data.
It seems an interesting viewpoint to consider the relationship between accumulation of mutations by in vitro passages during prolonged periods from first isolation in ATCC13950, and the difference of phenotypes between ATCC13950 and recently sampled clinical MAC-PD strains. However, there are no time-course samples of ATCC13950 isolates available. Therefore, we can neither investigate how many mutations have accumulated in a time-course manner, nor evaluate how much the accumulated mutations influence the phenotype in ATCC13950. It can be expected that the accumulation of in vitro mutations may cause the behavior of ATCC13950 different from clinical MAC-PD strains. However, it is to be elucidated yet which kinds of factors contribute to the characteristics of ATCC13950 that differ from clinical MAC-PD strains specifically.
It seems an interesting viewpoint to investigate the similarity of gene essentiality between genetical neighbor strains. However, we focused on the overview of the profiles of gene essentiality in clinical MAC-PD strains compared to ATCC13950. Thus, it was out of scope to elucidate the details of gene essentiality in each genetic phylogeny that clinical MAC-PD strains belong. The overview of phylogenetic trees should be referred to our previous publication on the comparative genomic analysis of 55 strains (Tateishi Y. BMC Microbiol. 2021; new Ref#5, new Supplementary Fig. 1), and we have shown Fig. 1 as the extracted phylogenetic tree of subject strains. To elucidate the details of gene essentiality in each genetic clade, it would be necessary to include a considerable number of strains that we used for comparative genomic analysis in 2021 (Tateishi Y. BMC Microbiol. 2021; new Ref#5). Furthermore, it would be necessary to set a referential control strain other than ATCC13950 for comparing gene essentiality. So far, it is not the highest priority for us to elucidate the similarity of gene essentiality between phylogenetic clades in detail, and such investigation will be planned as a future study.
The key features that separate ATCC13950 and clinical MAC-PD strains have not been proved yet, in contrast to the case of M. tuberculosis such as mutations in the gene of the response regulator PhoPR in the type strain H37Rv vs most clinical strains. However, the features that separate ATCC13950 and clinical MAC-PD strains may not be explained by a single genetic factor but may be explained by complicated factors such as epigenetic and/or regulatory factors. For example, the reason for the weakened virulence of H37Ra compared to H37Rv has not been able to be explained by simple genetic differences (Brosch R. Infect Immun. 1999).
In summary, we set the historical type strain ATCC13950 which is derived from infant abdominal lymphadenitis as a referential control strain for TnSeq analysis, because we intended to reveal the characteristics of clinical MAC-PD strains in terms of the gene essentiality and genetic requirements by comparing the clinical MAC-PD strains with the non-MAC-PD reference strain. We consider that the profiles of gene essentiality and genetic requirements specific to clinical MAC-PD strains confer the pathogenesis in an increasing number of MAC-PD patients worldwide without predisposing immunological disorders.
[References]
Cuttino, J.T. & Mc, C.A. Pure granulomatous nocardiosis, a new fungus disease distinguished by intracellular parasitism; a description of a new disease in man due to a hitherto undescribed organism, Nocardia intracellularis, n. sp., including a study of the biologic and pathogenic properties of this species. Am J Pathol 25, 1-47 (1949).
Kim, B.J. et al. Complete genome sequence of Mycobacterium intracellulare clinical strain MOTT-64, belonging to the INT1 genotype. J Bacteriol 194, 3268 (2012).
Carey, A.F. et al. TnSeq of Mycobacterium tuberculosis clinical isolates reveals strain-specific antibiotic liabilities. PLoS Pathog 14, e1006939 (2018).
Akusobi. C. et al.. Transposon-sequencing across multiple Mycobacterium abscessus isolates reveals significant functional genomic diversity among strains. mBio 6, e0337624 (2025).
Tateishi, Y. et al. Comparative genomic analysis of Mycobacterium intracellulare: implications for clinical taxonomic classification in pulmonary Mycobacterium avium-intracellulare complex disease. BMC Microbiol 21, 103 (2021).
Tateishi, Y. et al. Genome-wide identification of essential genes in Mycobacterium intracellulare by transposon sequencing - Implication for metabolic remodeling. Sci Rep 10, 5449 (2020)
Brosch R. et al. Genomic analysis reveals variation between Mycobacterium tuberculosis H37Rv and the attenuated M. tuberculosis H37Ra strain. Infect Immun. 67, 5768-74 (1999).
(2) Regarding the 'nine representative strains of M. intracellulare with diverse genotypes in this study,' how were these nine strains selected? To what extent do they represent the genetic diversity of the M. intracellulare population? A phylogenetic tree illustrating the global genetic diversity of the M. intracellulare population, with these strains marked on it, would be important to demonstrate their genetic representativeness.
Thank you for the comments on the selection of 9 subject strains. We selected the 9 strains based on the phylogenetic tree we published in 2021 (BMC Microbiol 2021; new Ref#5). We have shown the global phylogenetic tree of the M. intracellulare population in new supplementary Fig. 1. We have selected 4 or 5 strains from the major two groups (typical M. intracellulare group and M. paraintracellulare-M. indicus pranii group) for this TnSeq study, respectively.
[Reference]
Tateishi, Y. et al. Comparative genomic analysis of Mycobacterium intracellulare: implications for clinical taxonomic classification in pulmonary Mycobacterium avium-intracellulare complex disease. BMC Microbiol 21, 103 (2021).
(3) The authors observed a considerable amount of differential gene requirements in clinical strains. However, the genetic underpinning underlying the differential requirement of genes in clinical strains was not investigated or discussed. Because M. intracellulare has a huge number of accessory genes, the authors should at least check whether the differential requirement could be explained by the existence of a second copy of functional analogous genes or duplications.
Thank you for the comments on the effect of gene duplication on the change of genetic requirements between strains. Following the comments, we conducted blast search for the 162 genes showing increased Tn insertion reads in each subject strain. We found that M019 has duplicate genes of OCU_RS44705 coding adenosylhomocysteinase (LOCUS_42940: ahcY_1, LOCUS_21000: ahcY_2). However, there were no duplicate genes found in the remaining 161 genes showing increased Tn insertion reads.
From these results, we consider that gene duplication has minor effects on the change of genetic requirements between strains. Rather, sequence differences and accessory genes may play a key role in determining the difference of genetic requirements.
We have added a description of the above-mentioned result in the Result section (pages11-12, lines 191-199).
(4) Growth in aerobic and hypoxic conditions: The authors concluded that clinical strains are better adapted to hypoxia, as reflected by their earlier entry into the log phase. They presented the 'Time at midpoint' and 'Growth rate at midpoint.' However, after reviewing the growth curves, I noticed that ATCC13950 had a longer lag phase compared to other strains under hypoxic conditions, and its phylogenetic neighbor M018 also had a longer lag phase. Hence, I do not believe a conclusion can be drawn that clinical strains are better adapted to hypoxia, as this behavior could be specific to a particular clade. It's also possible that the ATCC13950 strain has adapted to aerobic growth. I would suggest that the authors include growth curves in the main figures. The difference in 'Time at midpoint' could be attributed to several factors, and visualizing the growth curves would provide additional context and clarity.
Thank you for the comments on the possibility of genotypes as a determinant of growth pattern in M. intracelulare. Following the comments, we performed aerobic and hypoxic growth assay in the two strains (M005 and M016) that neighbor ATCC13950.
Author response image 1.
The phylogenetic relationship between M005, M016 and ATCC13950. The former two strains are squared in blue.
M005 reached midpoint later than ATCC13950 both in aerobic and hypoxic conditions. By contrast, M016 reached midpoint three quarters earlier than ATCC13950 under hypoxic conditions. The growth rate was not significantly different between M005, M016 or ATCC13950 under either aerobic or hypoxic conditions, although P-value of M005 vs ATCC13950 was 0.0512 under aerobic conditions on Steel’s multiple comparisons test.
From the data of growth pattern in M005 and M016, we suggest that in addition to gene essentiality, genotypes may have some impact on the bacterial growth pattern under hypoxia; however, since there was a significant difference in the timing of hypoxic adaptation among ATCC13950 and its neighbor strains, bacterial growth pattern under hypoxia is considered to be determined by multiple factors such as genetic requirements and unproven regulatory systems. Taking into consideration that there are lots of genetically diverse strains other than ATCC13950 clade, many clinical MAC-PD strains are possibly better adapted to hypoxia.
Responding to the reviewer’s recommendation, we have added the description of the above-mentioned result in the revised manuscript (page 18, lines 313-322). And we have shown the data of growth curves of the original 9 subject strains in the new Fig 7. And we have added the data of the growth curves of M005 and M016 in new Supplementary Fig 7. Additionally, we have corrected the label of y-axis in new Fig. 7a and new Supplementary Fig. 7a and added the description as “Data are represented as CFUs in 4 μl sample at each timepoint.” in the Figure legends. (page 58, lines 1027-1028 and Supplementary Fig. 7a legend)
(5) Lack of statistical statement: The authors emphasized the role of pellicle-formation-associated genes in strain-dependent essential and accessory essential genes. Additionally, the authors observed that 10% of the genes required for mouse infection are also required for hypoxic pellicle formation. However, these are merely descriptive statements. There is no enrichment analysis to justify whether pellicle-formation-associated genes are significantly enriched in these groups.
Thank you for the comments on the enrichment of pellicle-formation associated genes in strain-dependent essential and accessory essential genes. We performed GSEA and found that 9.1% (16 out of 175) genes were hit as core enrichment. Of them, 4 genes were hit commonly as genes showing increased genetic requirements analyzed by resampling plus HMM analyses including genes of phosphoenolpyruvate carboxykinase pckA (OCU_RS48660), type VII secretion-associated serine protease mycP5 (OCU_RS38275), type VII secretion protein eccC5 (OCU_RS38345) and glycine cleavage system amino-methyltransferase gcvT (OCU_RS35955).
Author response image 2.
We have added the description of GSEA result in the revised manuscript (page 8, lines 137-144; Supplementary Fig. 2; Supplementary Table 5).
Reviewer #1 (Recommendations For The Authors):
Tn-seq and hypoxia adaption in clinical isolates of M. intracellulare (Mi): The authors claim that clinical strains are better adapted to hypoxia because their genetic requirements for optimum fitness overlap with genetic requirements for fitness of the type strain under hypoxia. This is a reasonable hypothesis, but it has not been well-supported by the data presented in Figure 7. The growth rates (Figure 7b) of most of the clinical strains under hypoxia appear to be less than the type strain, although they all seem to grow better than the type strain under normoxia. Perhaps a continuous growth curve of each strain, both as pure and mixed cultures under these conditions will provide a clearer picture.
Thank you for the comments on the difference of adaptation of hypoxic growth between ATCC13950 and MAC-PD strains. To clarify, growth rates shown in Figure 7 were calculated at the inflection point (midpoint) of the growth curves, which were modeled using a four-parameter logistic (4P logistic) model. As described in the Discussion, we found the pattern of hypoxic adaptation characteristics of the clinical MAC-PD strains from the growth curve forms. Taking into consideration the impact of growing bacteria on the disease progression of MAC-PD, the slow growth with early entry to log phase implicates the continuous impact on the infected hosts during logarithmic bacterial growth, which may be involved in the persistent and steadily progressive illness of MAC-PD for years in humans.
Unlike time-lapse imaging assay, the completely seamless sampling of cultures for CFU assay is impossible. Nevertheless, we collected sufficient timepoints to allow reliable curve fitting with the 4P logistic model, and thus consider our growth data to represent a valid approximation of continuous growth dynamics.
Regarding the suggestion of mixed culture experiments, we agree that such studies could be informative. However, co-culture conditions introduce additional variables, including inter-strain competition or synergy, which can obscure the specific contributions of hypoxic adaptation in each strain. Therefore, we believe that the current approach using monoculture growth curves under defined oxygen conditions offers a clearer interpretation of strain-specific hypoxic responses.
In vivo studies: It is unclear how virulent the two clinical strains, Mi27 and Mi198 are in the mouse model. The CFU data in Figure S1b reports the bacterial burden of the Tn libraries of the two strains, of which the overall population of Mi27 library seems to be declining. Without any information on the CFU, animal survival, and tissue pathology from the pure strains, data from the library will have limited implications.
Thank you for the comments on the data presentation of in vivo studies. We previously revealed the time-course data on CFUs, animal survival, and tissue pathology from the pure strains (Tateishi Y. BMC Microbiol. 2023; new Ref#22). Based on these data, we assumed that we would be able to harvest sufficient number of colonies from mice infected with M.i.27 or M.i.198, and we performed in vivo TnSeq studies using these two strains. We have referred to our previous publication on the virulence of MAC-PD pure strains used in this study for mice in the revised manuscript (page 12, line 212; new Ref #22).
The data of CFU counts were shown in new Supplementary Figure 3b. In the manuscript text, we explained as follows (page 12, lines 212-216): “The time course of the changes in the bacterial burden showed a pattern similar to those of the wild-type strains M.i.198 and M.i.27, respectively (Tateishi Y. BMC Microbiol. 2023; new Ref#22), except that it was not possible to harvest sufficient colonies (as few as 104/mouse) in the few mice infected with the M.i.27 Tn mutant strain in week 8 and week 16 (new Supplementary Fig, 3b, new Supplementary Table 8)”. The decline of overall population of M.i.27 Tn mutant library strains in the infected lungs can be explained by the lower virulence of M.i.27 pure strain that shows intermediate virulence phenotype than M.i.198 that shows high virulence phenotype.
[References]
Tateishi, Y. et al. Virulence of Mycobacterium intracellulare clinical strains in a mouse model of lung infection - role of neutrophilic inflammation in disease severity. BMC Microbiol 23, 94 (2023).
Data presentation: The manuscript suffers from a lack of clarity in data visualization and presentation, especially the Tn-Seq datasets. Panels describe the experimental workflow with a densely-worded paragraph, making it difficult to navigate through the major findings.
Thank you for the comments on the issue of Fig. 1b. Following the suggestion, we have modified the new Fig. 1b entitled “Strategy of the procedure of TnSeq analyses”.
Language: The paper should be extensively revised for language. Often the authors have mixed-up the terms like 'core' and 'accessory' 'genes' in lines 116-119 with 'core and accessory genomes' in Figure 2c, which is not even mentioned in the paper. It is further unclear how they identified 3153 and 5824 core and accessory genes, respectively, from 55 different strains of Mi. Line 251: ..."involved by confer..." needs revision. The terms "increased gene essentiality" and 'growth-defected associated genes" are very confusing. The essentiality of a gene is either absolute or conditional but is not quantitative. Similarly, 'growth-defect associated' can be replaced with a better phrase that alludes to fitness loss in the clone. Additional typos were found throughout the paper that need to be fixed.
Thank you for the comments on the issue of scientific words including “'core and accessory genomes” and “gene essentiality” used in this study.
Based on Rosconi’s paper (Panel C of Fig. 1 in Nat Microbiol. 2022; new Ref#14), we used the phrases “accessory genome and core genome” as a meaning of a whole set of genes belonging to accessory and core genes. To avoid the confusion and keep consistency, we replaced the term “genomes” to “genes” in the revised manuscript.
In our previous comparative genomic analysis, we identified 3153 and 5824 core and accessory genes, respectively, from 55 different strains of M. intracellulare (Tateishi Y. BMC Microbiol. 2021; new Ref #5). To perform pangenomic analysis, we used the software Bacterial Pan-Genome Analysis tool (BGAP) (Narendrakumar NM. Sci Rep 2016).
We admit that gene essentiality is a qualitative but not a quantitative trait. We have corrected the term "increased gene essentiality" as "increased genetic requirements" throughout the manuscript.
We have used the term “growth-defect (GD)” based on the classification of gene essentiality calculated by the Hidden Markov Model (HMM) complied by TRANSIT software (DeJesus. PLoS Comput Biol. 2015; new Ref#12). The HMM classifies genes as essential (ES), GD, non-essential (NE), growth-advantage (GA). GD means difficulties of growth (growth deficiency) in aerobic conditions in vitro, because Tn insertions are less frequent. The suggested phrases “fitness loss” or “less fit” may include the meaning of the comparison of two different conditions such as culture conditions exposed to a single bacterial strain. Since the HMM analysis is performed in data of a single strain in a specific bacterial condition, we consider that the phrase including “fitness” is somewhat unsuitable for describing the classification of gene essentiality. Thus, it is difficult for us to rephrase GD to the word that implies fitness levels between two conditions in a single bacterial strain.
[References]
Rosconi, F. et al. A bacterial pan-genome makes gene essentiality strain-dependent and evolvable. Nat Microbiol 7, 1580–1592 (2022).
Tateishi, Y. et al. Comparative genomic analysis of Mycobacterium intracellulare: implications for clinical taxonomic classification in pulmonary Mycobacterium avium-intracellulare complex disease. BMC Microbiol 21, 103 (2021).
Narendrakumar NM et al. BPGA- an ultra-fast pan-genome analysis pipeline. Sci Rep 2016 6, 24373 (2016).
DeJesus, M.A. et al. TRANSIT--A Software Tool for Himar1 TnSeq Analysis. PLoS Comput Biol 11, e1004401 (2015).
Reviewer #2 (Recommendations For The Authors):
Major Comments:
(1) Result 1 (Page 6-7): Common essential and growth-defect-associated genes representing the genomic diversity of M. intracellulare strains.
(1a) From Table S1, it is observed that the numbers of Tn-inserted TA sites significantly vary (p >0.05) among biological replicates for each strain when compared with the reference strain ATCC13950. the authors should provide an explanation of how they overcame this variation in their analysis.
Thank you for the comment on the issue of a variable number of Tn-inserted TA sites among biological replicates for each strain of MAC-PD.
On TRANSIT software, we set the replicate option as Sum to combine read counts. And we used Beta-Geometric correction (BGC) to normalize the datasets to fit an “ideal” geometric distribution with a variable probability parameter ρ.
Following the comment, we have added the description on which option we used for handling the replicate data and normalization (page 36, lines 640-643).
(1b) Importantly, saturation in most of the strains is only ~50-60%. In such a case, there will be a high probability that Tn will not hit a nonessential region due to chance instead of selection (See DeJasus et al., mBio, 2017). It has been observed that the sequence pattern (GC)GNTANC(GC) is strongly associated with non-permissiveness. As shown earlier, the authors need to carefully look for the potential non-permissive sites before concluding the fate of a gene. Also, they should acknowledge the potential limitations of their approach due to the suboptimal level of saturation.
Thank you for the comment on the saturation of Tn mutant libraries. Our method of comparison of genetic requirements between strains are the same as a previous report that used duplicate Tn mutant libraries of clinical Mtb strains of different genotypes and triplicate Tn mutant libraries of H37Rv for identifying increased genetic requirements of clinical Mtb strains (Carey. PLoS Pathog 2018; new Ref#8). Our method is also based on the coauthor’s TnSeq study on H37Rv (Minato Y. mSystems 2019; new Ref#61). Moreover, by combining replicates, the saturation of our Tn mutant libraries became 62-79% as follows: ATCC13950: 67.6%, M001: 72.9%, M003: 63.0%, M018: 62.4%, M019: 74.5%, M.i.27: 76.6%, M.i.198: 68.0%, MOTT64: 77.6%, M021: 79.9%. That is, we calculated gene essentiality from the Tn mutant libraries with 62-79% saturation in each strain. The levels of saturation of transposon libraries in our study is similar to the very recent TnSeq anlaysis by Akusobi where 52-80% saturation libraries (so-called “high-density” transposon libraries) are used for HMM and resampling analyses (Supplemental Methods Table 1[merged saturation] in Akusobi. mBio. 2025; new Ref#9). The saturation of Tn insertion in individual replicates of our libraries is also comparable to that reported by DeJesus (Table S1 in mBio 2017; new Ref#57). Thus, we consider that our TnSeq method of identifying essential genes and detecting the difference of genetic requirements between clinical MAC-PD strains and ATCC13950 is acceptable.
As the Reviewer indicates, there is non-permissive sequence pattern in mariner transposon mutagenesis. Using more than 10 replicates of Tn mutant libraries is quite an accurate method for detecting essential genes in nonstructural small genes such as small regulatory RNAs. However, as DeJesus shows, the number essential genes identified by TnSeq are comparable in large genes possessing more than 10 TA sites between 2 and 14 TnSeq datasets, most of which seem to be structural genes (Supplementary Fig 2 in mBio 2017; new Ref#57). Thus, we do not consider that we made a serious mistake for the classification of essentiality in most of the structural genes that encode proteins. With respect to the coverage of non-permissive sites, our TnSeq method might need to be improved if it is intended to classify the gene essentiality quite accurately on the small genes including small regulatory RNAs.
We investigated the non-permissive TA sites in ATCC13950. There are 4136 (6.43% of total ORFs) nonpermissive TA sites in ATCC13950, which is less than in H37Rv (9% of total ORFs) (DeJesus MA. mBio 20171; new Ref#57) and in M. abscessus ATCC19977 (8.1% of total ORFs)(Rifat D. mBio. 2021; new Ref#58). As for larger ORFs (TA sites > = 10), there are nonpermissive TA sites in 89 genes (ORFs) of common “essential (ES)” or “growth-defect-associated (GD)” (4.82% of a total of 1844 larger ORFs in ATCC13950). As for small ORFs (2-9 TA sites), there are nonpermissive TA sites in 41 genes (ORFs) of common ES or GD (1.35% of a total of 3021 smaller ORFs in ATCC13950).
We appreciate the idea of concluding the fate of gene essentiality by the presence/absence of non-permissive TA sites. However, we cannot conclude the fate of gene essentiality classification only by the presence/absence of potential non-permissive sites. Because, strictly to say, it is impossible to conclude the scientific truth of gene essentiality without functional analysis using gene manipulation. In accurate, TnSeq can “predict” the gene essentiality but cannot perfectly guarantee the functional significance. However, in the current situation, most of the recent TnSeq studies have been published only by the TnSeq analysis without functional analysis that uses gene manipulation strains of all targets they identified. Taking such limitations of TnSeq including non-permissive sites into consideration, we consider that the essentiality of the detected genes should be determined in further studies, mainly including biological experiments such as functional studies using gene manipulation strains.
We have added the above-mentioned contents in the revised manuscript (pages 32-33, lines 559-580).
[References]
Carey, A.F. et al. TnSeq of Mycobacterium tuberculosis clinical isolates reveals strain-specific antibiotic liabilities. PLoS Pathog 14, e1006939 (2018).
Minato, Y., et al. Genomewide assessment of Mycobacterium tuberculosis conditionally essential metabolic pathways. mSystems. 4, e00070-192019 (2019).
Akusobi. C. et al. Transposon-sequencing across multiple Mycobacterium abscessus isolates reveals significant functional genomic diversity among strains. mBio 6, e0337624 (2025).
DeJesus, M.A. et al. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio 8, e02133-16 (2017).
Rifat, D., Chen L., Kreiswirth, B.N. & Nuermberger, E.L.. Genome-wide essentiality analysis of Mycobacterium abscessus by saturated transposon mutagenesis and deep sequencing. mBio 12, e0104921 (2021).
(1c) Line 100: Authors report a total of 131 genes identified as essential or growth-defect-associated with the HMM analysis across all M. intracellulare strains. It should be explained in more detail how gene essentiality was determined (see above comment in (1b)). Furthermore, in Table S3 authors should mention the essential and growth defective trait of each of the 131 genes.
Thank you for the comment on how to classify the 131 genes as essential or growth-defect-associated with the HMM analysis across all M. intracellulare strains. As replied in (1b), the average saturation of Tn insertion of our libraries became 62-79% when combining duplicate or triplicate data in each strain. The levels of saturation of transposon libraries in our study is similar to the very recent TnSeq analysis by Akusobi where 52-80% saturation libraries (so-called “high-density” transposon libraries) were used for HMM and resampling analyses, and most of triplicate libraries ranges 70-79% saturation (Supplemental Methods Table 1[merged saturation] in Akusobi. mBio. 2025; new Ref#9). The saturation of Tn insertion in individual replicates of our libraries is also comparable to those with DeJesus (Table S1 in mBio 2017; new Ref#57). Thus, we consider that our TnSeq libraries are acceptable for identifying essential genes and growth-defect-associated genes by the HMM method.
We used the HMM method as reported by DeJesus (DeJesus. PLoS Comput Biol. 2015; new Ref#12). HMM method can categorize the gene essentiality throughout the genome including “Essential”, “Growth-defect”, “Non-essential” and “Growth-advantage”. “Essential” genes are defined as no insertions in all or most of their TA sites. “Non-essential” genes are defined as regions that have usual read counts. “Growth-defect” genes are defined as regions that have unusually low read counts. “Growth-advantage” genes are defined as regions that have unusually high low read counts.
Following the previous report (Carey AF. PLos Pathog 2018; new Ref#8), the annotation for the clinical MAC-PD strains was adapted from that of ATCC13950 by adjusting the START and END coordinates of each ORF in the clinical MAC-PD strains according to their alignment with the corresponding ORFs of ATCC13950. By using an adjusted annotation table, gene essentiality was classified by the HMM analysis.
We have added the explanation of how we identified essential and growth-defect-associated genes in the Methods (pages 35-36, lines 620-632). And following the comment, we have added the data of classification of gene essentiality in the 131 genes in the new Supplementary Table 3 in the revised manuscript.
[Reference]
DeJesus, M.A. et al. TRANSIT--A Software Tool for Himar1 TnSeq Analysis. PLoS Comput Biol 11, e1004401 (2015).
Carey, A.F. et al. TnSeq of Mycobacterium tuberculosis clinical isolates reveals strain-specific antibiotic liabilities. PLoS Pathog 14, e1006939 (2018).
Akusobi. C. et al. Transposon-sequencing across multiple Mycobacterium abscessus isolates reveals significant functional genomic diversity among strains. mBio 6, e0337624 (2025).
DeJesus, M.A. et al. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio 8, e02133-16 (2017).
(1d) In Table S4, the authors show strain-specific putative essential genes from the core and accessory gene sets. For the sake of clarity, it is important to have the name of all the strains against each gene in which it is predicted essential or growth defective.
Thank you for the comment on the hit strains on the genes classified as strain-specific and accessory putative essential of growth-defect associated. Following the comment, we have added the data of hit strains in the new Supplementary Table 4 in the revised manuscript.
(1e) Lines 123-126: It is not clear what is the relevance of highlighting genes involved in hypoxic pellicle formation in ATCC13950. These appear to be randomly distributed across different clinical isolates and is not clear whether they correlate with differential susceptibility of the reference strain and clinical isolates to hypoxia.
Thank you for the comment on the relevance of highlighting genes involved in hypoxic pellicle formation in ATCC13950. The rationale for the importance of hypoxic pellicle genes in clinical MAC-PD strains is that the profiles of genetic requirements in each bacterial strain reflect the adaptation to the environment in which each strain lives. When the strains are placed in a special environment, they can adapt to the situation by altering the profiles of genetic requirements, resulting in the remodeling of metabolic pathways. We indeed found that the genetic requirements of several hypoxic pellicle genes were increased in clinical MAC-PD strains in vitro situations. These data suggest the hypoxic pellicle genes become more important in clinical MAC-PD strains for in vitro growth than in ATCC13950.
Moreover, hypoxia is known to be one of the characteristic conditions in vivo including clinical lesions (McKeown. Br Br J Radiol. 2014). We consider it reasonable to expect that the strains derived from MAC-PD patients without predisposing immunological disorders may adapt under hypoxic conditions for maintaining bacterial survival. Therefore, we highlighted the genes involved in hypoxic pellicle formation in ATCC13950.
We have added the description of the rationale for the importance of hypoxic pellicle genes in clinical MAC-PD strains in the revised manuscript (page 9, lines 148-155).
[Reference]McKeown, S.R., et al. Defining normoxia, physoxia and hypoxia in tumours-implications for treatment response. Br Br J Radiol 87,: 20130676 (2014).
(2) Result 2 (pages 8-10): Genes with increased gene essentiality in clinical MAC-PD strains are also required for hypoxic pellicle formation in the type strain.
(2a) As reported by authors (lines 123-126), only a small fraction of genes showing essentiality in clinical MAC-PD strains are required for hypoxic pellicle formation in the reference strain, which might be due to random distribution. Authors should avoid making such a generalised statement that reflects the association of the entire essential gene pool in clinical MAC-PD strains with hypoxic pellicle formation.
Thank you for the comment on the issue of a small fraction of genes showing increased genetic requirements in clinical MAC-PD strains that is shared with genes required for hypoxic pellicle formation in the type strain ATCC13950. We admit that the section title may mislead that the genes required for hypoxic pellicle formation confer the entire essential gene pool of clinical MAC-PD strains. Following the comment, we have revised the section title as “Partial overlap of the genes showing increased genetic requirements in clinical MAC-PD strains with those required for hypoxic pellicle formation in ATCC13950” (page 9, lines 146-147).
We consider that it cannot be explained by a mere coincidence that we obtained the data of partial overlap of genes showing essentiality in clinical MAC-PD strains with genes required for hypoxic pellicle formation in ATCC13950, because we demonstrated the supporting data such as the pattern of genetic requirements suggesting gluconeogenic metabolic shift (Fig. 5) and the different pattern of hypoxic growth curves between clinical MAC-PD strains and ATCC13950 (Fig. 7).
(2b) I fail to understand how the number of Tn insertions determines "more" or "less" essentiality of a gene particularly with 50-60% saturation. To my understanding, essentiality is a qualitative trait. Either a gene will be essential (based on no Tn insertion despite having the permissive sites), critical (poor representation of Tn insertions at the permissive sites due to growth defect of the strain in the pool), non-essential (expected frequency of insertion) or growth-advantageous (higher representation of Tn insertions at the permissive sites due to growth advantage of the strain in the pool). Hence, authors should avoid quantifying the essentiality of a gene.
Thank you for the comments on the trait of gene essentiality. We realize that essentiality is a qualitative trait, not a quantitative trait. Taking into consideration the number of Tn insertions determines "more" or "less" requirements of a gene, we have corrected the manuscript by using the phrase “genetic requirements” instead of “gene essentiality”.
As mentioned earlier, our method of comparison of genetic requirements between strains are the same as a previous report that used duplicate Tn mutant libraries of clinical Mtb strains of different genotypes and triplicate Tn mutant libraries of H37Rv for identifying increased genetic requirements of clinical Mtb strains (Carey AF. PLoS Pathog 2018; new Ref#8). Moreover, as described in rebuttal (1b), the saturation of our Tn mutant libraries by combining replicates are 62-79% as follows: ATCC13950: 67.6%, M001: 72.9%, M003: 63.0%, M018: 62.4%, M019: 74.5%, M.i.27: 76.6%, M.i.198: 68.0%, MOTT64: 77.6%, M021: 79.9%. That is, we calculated gene essentiality from the Tn mutant libraries with 62-79% saturation in each strain. The levels of saturation of transposon libraries in our study is similar to the recent TnSeq analysis by Akusobi where 52-80% saturation libraries (“high-density” transposon libraries) were used for HMM and resampling analyses (Supplemental Methods Table 1[merged saturation] in Akusobi C. mBio. 2025; new Ref#9).
Thus, we consider that our data of the difference of genetic requirements between clinical MAC-PD strains and ATCC13950 are acceptable.
[Reference]
Akusobi. C. et al. Transposon-sequencing across multiple Mycobacterium abscessus isolates reveals significant functional genomic diversity among strains. mBio 6, e0337624 (2025).
(2c) From Figures 3-4, it seems the authors intend to highlight the insertion frequencies of certain genes in the clinical isolates compared to those in the reference strain to conclude whether a gene has become more critical and its disruption results in the growth defective phenotype (poor representation) in the clinical isolates, or a critical/essential gene has become dispensable in these strains.
Based on these arguments, I suggest that the authors modify the title of the result such as "Tn insertion reveals differential requirement of genes for in vitro growth of clinical MAC-PD strains" or "Identification of genes differentially required for in vitro growth of clinical MAC-PD strains" as this is precisely the information we gain from this section of the study. Also, it is suggested to re-draft the rationale of this section as only 4 genes associated with hypoxic pellicle formation, were found to exhibit reduced insertion frequencies in the clinical isolates out of total of 283 genes. Hypoxia-related genes can be highlighted in the next section (see below).
Thank you for the suggestion to modify the section title and to re-draft the rationale of the section. Following the comment, we modified the section title as “Partial overlap of the genes showing increased genetic requirements in clinical MAC-PD strains with those required for hypoxic pellicle formation in ATCC13950 (page 9, lines 146-147)
Following the suggestion, we have revised the rationale of this section as follows: “The sharing of strain-dependent and accessory essential or growth-defect-associated genes with genes required for hypoxic pellicle formation in ATCC13950 prompts us to consider that the profiles of gene essentiality in clinical MAC-PD strains may be associated with the genes required for hypoxic pellicle formation in ATCC13950.” (page 9, lines 151-155)
The reviewer points out that only 4 genes associated with hypoxic pellicle formation were found to exhibit reduced insertion frequencies in the clinical isolates out of total of 283 genes. However, to discuss how much proportion of the genes were detected to be increasingly required in clinical MAC-PD strains compared to ATCC13950, we should focus on the 121 genes showing increased requirements in clinical MAC-PD strains compared to ATCC13950, excluding the 162 genes indispensable for clinical MAC-PD strains. Thus, we described that 4 genes associated with hypoxic pellicle formation, were found to exhibit reduced insertion frequencies in the clinical isolates out of the 121 genes having significantly fewer Tn insertions than ATCC13950 in the manuscript (Fig. 3).
(3) Result 3 (Page 10-14): Requirement of genes with increased gene essentiality in the clinical MAC-PD strains for mouse lung infection.
(3a) The title should be modified to "Identification of genes in the clinical MAC-PD strains required for mouse lung infection".
Following the comment, we have modified the section title as "Identification of genes in the clinical MAC-PD strains required for mouse lung infection". (page 12, lines 201-202).
(3b) Further, the rationale of this experiment needs to be modified. As mentioned above, up until now the impact of hypoxic pellicle formation genes in the growth of MAC-PD strains remains unconvincing. The rationale of mouse infection experiments could be straightforward- "to identify genes critical for animal infection of the clinical isolates".
Thank you for the comment on the rationale of the in vivo TnSeq experiment. Following the comment, we have revised the rationale as “The impact of hypoxia on mycobacteria under various ecological circumstances implies that the genes required for pathogenesis of MAC-PD may be in some degrees, overlapped with the genes with increased requirements in the clinical MAC-PD strains compared to ATCC13950, and also with the genes required for hypoxic pellicle formation in ATCC13950. To identify genes required for in vivo infection of clinical MAC-PD strains,” in the revised manuscript (page 12, lines 204-210).
(3c) The authors should avoid using the term "genes with increased essentiality" for the reasons explained above in point #2b.
Following the comment, we have corrected the term as “genes with increased requirements” in the revised manuscript (page 12, line 207).
(3d) From Tables S8 and S9, I can find 93 genes in Mi198Tn and 74 genes in Mi27Tn for which Tn insertion mutants are under-represented in TnSeq at all time points from Day 1 to Wk 16 in comparison to input. Importantly, excluding results from Day 1 when the infection has just settled, I find 172 and 121 genes in Mi198Tn and Mi27Tn, respectively, under-represented in lungs between Wk 4-16. My suggestion is that authors should focus more on such genes and identify the characteristics of these genes and what fraction belongs to those involved in hypoxic pellicle formation in the reference strain. I am perplexed why authors have categorically ignored other genes and only focused on a set of genes that correspond to ~10-12% of entire differentially abundant mutant pool.
Thank you for the suggestion on the genes that Tn insertion mutants are under-represented in TnSeq from Weeks 4-16 in the infected mouse lungs be analyzed for overlapping the genes involved in hypoxic pellicle formation in the type strain ATCC13950. We found that at all timepoints from Day1 to Week 16, 74 genes and 99 genes were under-represented in lungs infected with M.i.27Tn and M.i.198Tn, respectively. Of them, 21 (28.3%) and 12 (12.1%) genes belonged to the genes involved in the genes required for hypoxic pellicle formation in the type strain. We found that at timepoints from Week 4 to Week 16, 121 genes and 172 genes were under-represented in lungs infected with M.i.27Tn and M.i.198Tn, respectively. Of them, 21 (23.1%) and 30 (18.0%) genes belonged to genes involved in hypoxic pellicle formation in the type strain. These hypoxic pellicle-associated genes detected both in M.i.27 and M.i.198 encoded methionine synthesis, acyl-CoA dehydrogenase, isocitrate lyase, MMPL family transporter at all time points (from Day1 to Week 16). And additionally, multifunctional oxoglutarate decarboxylase/dehydrogenase, proteasome subunits, ABC transporter ATP-binding protein/permease, lipase chaperone at all time points (from Week 4 to Week 16). We have described these results in the Result section (page 14 lines 236-248) and new Supplementary Tables 12 and 13.
As for M. intracellulare, conditionally essential genes have not been revealed except for those required for hypoxic pellicle formation in ATCC13950 revealed by us (Tateishi Y. Sci Rep. 2020; new Ref#10). This study is the first to focus on the relationship between the difference of genetic requirements among strains and hypoxic adaptation. We found a certain proportion of overlapped genes required for mouse lung infection and ATCC13950’s hypoxic pellicle formation. We consider it reasonable to focus on the category of genes required for hypoxic pellicle formation to analyze the datasets of TnSeq in mice.
[Reference]
Tateishi, Y. et al. Genome-wide identification of essential genes in Mycobacterium intracellulare by transposon sequencing - Implication for metabolic remodeling. Sci Rep 10, 5449 (2020).
(3e) Page 13, lines 224-227: "Despite the differences in the profiles of the genes required for in vivo infection between strains, these data suggest that increased gene essentiality for hypoxic growth confers advantages for pathogenesis in vivo."
For the reason described above, I find it a misleading hypothesis that hypoxic growth confers advantages for pathogenesis in vivo. How come only 10-12% of the entire gene sets which include genes of varying functions, can be the sole contributors to bacterial survival in host organelles during infection?
More importantly, the mouse is not considered a good model for hypoxia as mouse infection does not lead to the formation of solid granuloma with a hypoxic core Though I am not convinced with the authors' bias toward hypoxia-related genes, however, if at all they aim to investigate the role of such genes by an unbiased enrichment of TnSeq mutant, they should have used C3HeJ mice which are known to form granulomas (Boute et al., 2017 (doi: 10.1186/s13567-017-0477-7)).
Thank you for the comments on the issue of the contribution of genes required for hypoxic growth and on the difference of hypoxic levels between mouse lineages. We did not intend to mention that a set of genes required for hypoxic growth is the sole contributor to bacterial survival in host organs during infection. As we discussed in the Discussion section, we acknowledge that the adaptation to the difference of carbon source between _in vitr_o and in vivo infection (i.e. preferential usage of lipid carbon source in vivo) is involved in the pathogenesis of mycobacterial diseases (Yang. Front Microbiol 2018; new Ref#33, Gouzy. Proc Natl Acad Sci U S A 2021; new Ref#29, Quinonez. mBio 2022; new Ref#40, Pandey. Proc Natl Acad Sci U S A 2008; new Ref#41). We consider that not only the genes required for hypoxic pellicle formation but also strain-dependent/accessory genes conferring kinds of metabolism other than hypoxic pellicle formation can be estimated to be involved in the in vivo mouse lung infection.
We have modified the sentence to clearly express our intention as follows: “These in vivo TnSeq data suggest that, despite the differences in the profiles of the genes required for in vivo infection between strains, increase of genetic requirements for hypoxic growth in part contribute to the pathogenesis in vivo.” (pages 15-16, lines 269-271)
It seems to be an interesting idea to perform TnSeq by using C3HeJ mice. The granuloma formed in C3HeJ mice becomes extremely hypoxic (less than 1%, corresponding the level of “pathological” hypoxia) which is as severe as the detection range by pimonidazole. In our model, the effect of such pathological levels of hypoxia on granuloma formation might not be detected. However, the lesion formed in C57BL/6 mice becomes a “physiological” level of hypoxia (5% O2) (McKeown SR. Br Br J Radiol. 2014) which is the same O2 level for M. intracellulare to form pellicles. In principle, oxygen levels inside human bodies are physiologically hypoxic, and many biological events are experimentally investigated in this condition. Thus, we consider that we were able to observe the effect of physiological hypoxia on M. intracellulre growth both in vitro (hypoxic pellicles) and in vivo (infected C57BL/6 mice).
[Reference]
Yang, T. et al. Pan-genomic study of Mycobacterium tuberculosis reflecting the primary/secondary genes, generality/individuality, and the interconversion through copy number variations. Front Microbiol 9, 1886 (2018).
Gouzy, A., Healy, C., Black, K.A., Rhee, K.Y. & Ehrt, S. Growth of Mycobacterium tuberculosis at acidic pH depends on lipid assimilation and is accompanied by reduced GAPDH activity. Proc Natl Acad Sci U S A 118, e2024571118 (2021).
Quinonez, C.G. et al. The role of fatty acid metabolism in drug tolerance of Mycobacterium tuberculosis. mBio 13, e0355921 (2022).
Pandey, A.K. & Sassetti, C.M. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci U S A 105, 4376-4380 (2008).
McKeown., S.R. et al. Defining normoxia, physoxia and hypoxia in tumours-implications for treatment response. Br Br J Radiol 87, 20130676 (2014).
(3f) An important set of data with the ATCC13950 reference strain is missing here. It is suggested that authors perform this study with the reference strain to identify whether the enrichment of genes is similar across all strains or is specific to the clinical isolates.
Thank you for the comment on the setting of ATCC13950 as a control strain in the mouse infection experiment. However, we proved that bacterial burden of ATCC13950 is reduced continuously from 4 weeks of infection, and that ATCC13950 is almost completely eliminated from 8 to 16 weeks of infection (BMC Microiol 2023; new Ref#22). Therefore, it is impossible to perform TnSeq to detect the genes required for persistent infection in mice infected with ATCC13950.
[Reference]
Tateishi, Y. et al. Virulence of Mycobacterium intracellulare clinical strains in a mouse model of lung infection - role of neutrophilic inflammation in disease severity. BMC Microbiol 23, 94 (2023).
(3g) Pages 13-14, lines 228-245: "We have performed a statistical enrichment analysis of gene sets by GSEA...".
The comparison made here is not clear to me. It seems the authors do compare genes required for the growth of M.i.27 and M.i.198 in mouse lungs with the gene sets required for hypoxic pellicle formation in ATCC13950 together with the gene sets showing increased gene essentiality observed in the clinical MAC-PD strains, and claim that a significant % of genes belong to hypoxia-adaptation pathways. It is factually incorrect because a majority of these might overlap with those found critical for the in vitro survival of MAC-PD strains. It is suggested that authors re-analyze their data by comparing genes required for the growth of M.i.27 and M.i.198 in mouse lungs individually with those involved in hypoxic pellicle formation in ATCC13950, and with the gene sets found critical for in vitro growth, and present accordingly.
Thank you for the suggestion on the re-analysis of gene enrichment analysis of genes required for M.i.27 and M.i.198 in vivo infection, individually with genes involved in hypoxic pellicle formation in ATCC13950 and with those showing genetic requirements in clinical MAC-PD strains compared to ATCC13950.
About 50% (92 and 94 out of 181 genes through Day 1 to Week 16 and through Week4 to Week16 of infection) and 40% (70 and 79 out of 179 genes through Day 1 to Week 16 and through Week 4 to Week 16 of infection) of genes required for hypoxic pellicle formation in ATCC13950 were listed as enriched in genes required for mouse lung infection in M.i.27 and M.i.198, respectively. In addition, about 42% (54 and 56 out of 128 genes through Day 1 to Week 16 and through Week 4 to Week 16 of infection) and 40% (79 and 68 out of 179 genes through Day 1 to Week 16 and through Week 4 to Week 16 of infection) of genes showing increased requirements in clinical MAC-PD strains compared to ATCC13950 were listed as enriched in genes required for mouse lung infection in M.i.27 and M.i.198, respectively.
The tables and graphs of GSEA results are shown in Supplementary Figs. 5, 6.
These data indicate that 40-50% of the genes required for in vitro hypoxic pellicle formation and the strain-dependent/accessory essential genes are significantly enriched individually with in vivo bacterial growth. We have added the result of reanalyzed data of GSEA in the Result (pages 16-17, lines 287-290). We have shown the detail of reanalyzed data of GSEA in Supplementary Figs. 5, 6 and Supplementary Tables 15, 16.
(3h) Since authors have used Tnseq of pooled mutants, which often yields misleading information, it is important to validate some of their findings upon mouse infection with individual mutants that yield prominent as well as baseline reduction at different time points. In the absence of validation, it remains a mere speculation of the role of these genes in the infection of these strains to animals.
Thank you for the suggestion on the validation of the TnSeq hit genes on the in vivo survival. We acknowledge the importance of validating the TnSeq-hit genes by constructing knockout mutants. We have recently succeeded in constructing the vectors for making knockout strains of M. intracellulare (Tateishi Y. Microbiol Immunol. 2024). We will proceed to the infection experiment of knockout mutants by using our system for constructing them.
[Reference]
Tateishi Y. et al. Construction of knockout mutants in Mycobacterium intracellulare ATCC13950 strain using a thermosensitive plasmid containing negative selection marker rpsL+. Microbiol Immunol 68, 339-347 (2024).
(4) Result 4 (Page 14-15): Preferential hypoxic adaptation of clinical MAC-PD strains evaluated with bacterial growth kinetics.
(4a) "The metabolic remodeling, such as the increased gene essentiality of gluconeogenesis and the type VII secretion system..". As stated above, the essentiality of a gene, being a qualitative trait, should not be presented in quantitative terms. The authors should re-phrase this statement.
Following the comment, we have corrected the term as “The metabolic remodeling, such as the increased genetic requirements of gluconeogenesis and the type VII secretion system.” (page 17, lines 296-297)
(4b) "overlap of the genes required for mouse lung infection and those required for hypoxic pellicle formation involved by conferring these metabolic pathways..". There is a syntax error in this statement and needs revision.
Following the comment, we have corrected the phrase as “overlap of the genes required for mouse lung infection and those required for hypoxic pellicle formation involved by these metabolic pathways”. (page 17, lines 297-299)
(4c) The altered requirement of genes in different clinical strains for survival provides only circumstantial evidence of metabolic remodeling. Authors are suggested to perform metabolic profiling of representative clinical and reference strains, as it is important to examine whether these bacteria indeed undergo metabolic shift.
Thank you for the comment on the metabolic profiling of the representative clinical and reference strains. We previously published the TnSeq result of ATCC13950 and we produced the current data by organizing with our previous findings (Fig. 4 in Tateishi Y. Sci Rep 2020; new Ref#10). The priority of the current study was to elucidate the difference and diversity of genetic requirements between clinical MAC-PD strains and ATCC13950. We consider that it is of some value to show the even circumstantial evidence of metabolic remodeling by TnSeq, because it provides a strong rationale for proceeding to the next study including metabolomic analysis.
[Reference]
Tateishi, Y. et al. Genome-wide identification of essential genes in Mycobacterium intracellulare by transposon sequencing - Implication for metabolic remodeling. Sci Rep 10, 5449 (2020).
(5) Result 5 (Page 16-18): Effects of knockdown of universal and accessory/strain-dependent essential or growth-defect-associate genes in clinical MAC-PD strains.
(5a) Lines 273-277: The rationale of using CRISPRi should be correctly presented to evaluate the effect of individual genes' suppression on the downstream phenotype and not to establish the CRISPRi silencing tool in MAC.
Thank you for the comment on the rationale of the section of the CRISPR-i experiment. Following the comment, we have modified the sentence as follows: “With an intention to evaluate the effect of suppressing TnSeq-hit genes on bacterial growth.” (page 19, lines 333-334 in the revised manuscript).
(5b) Line 278: pRH2052/pRH2521 are the plasmids and not the CRISPRi system.
Following the comment, we have corrected the phrase as “pRH2052/pRH2521 clustered regularly interspaced short palindromic repeats interference (CRISPR-i) plasmids.” (page 19, lines 334-335 in the revised manuscript).
(5c) Line 280: Other pioneering studies on the use of CRISPRi for gene silencing in mycobacteria (Chaudhary et al., Nat Comm, Rock et al., Nat Microbio) should also be cited.
Thank you for the comment on adding the reference papers on CRISPR-i in mycobacteria. We have added the two suggested papers in the revised manuscript as new Ref #30 and #31. (page 19, line 336).
(5d) Lines 282-283: It is not clear why M001 and MOTT64 could not be transformed. Did the authors use any control plasmid to evaluate the transformation efficiency of these strains?
Thank you for the comment on the failure of transformation in M001 and MOTT64.
Following the comment, we have performed the experiment for evaluating the efficiency of transformation in the 9 M. intracellulare strains we used in this study. We have used an E. coli-mycobacteria shuttle vector pSO246KM-Prhsp65-luc that expresses firefly luciferase as a control plasmid (Aoki K. J Biol Chem 2004). For obtaining transformed colonies, we used 7H10/OADC agar plates containing the same concentration of kanamycin that we used for preparing Tn mutant libraries and for obtaining CRSISPR-i knockdown strains.
We have observed no colonies grown on agar plates in MOTT64 after electroporation of the pSO246KM-Prhsp65-luc plasmid. In most of the remaining strains, the transformed colonies have emerged fully on day 10 of culture after electroporation of the plasmid. However, we have observed that M001 needs twice as long as a period for the emergence of transformed colonies. On day 21, the number of colonies in M 001 have finally become comparable to that of the other strains. We have checked the luciferase activity of 6-12 colonies in each strain except for MOTT64, and we have confirmed the transformation of the plasmid by the data of higher luciferase activity in the colonies undergoing electroporation of the plasmid than in those not undergoing electroporation.
The possible reason for the incapability of obtaining transformants of CRISPR-i vectors in MOTT64 may be due to the extremely low efficiency of acquiring foreign DNA. And the possible reason for the incapability of obtaining transformants of CRISPR-i vectors in M001 may be intolerable to the stress caused by transformation of plasmids compared to other M. intracellulare strains. For M001, pSO246KM-Prhsp65-luc plasmid may cause tolerable stress for transformation, resulting in the delayed emergence of transformed colonies. By contrast, the CRIPSR-i plasmids may cause greater stress for M001 than pSO246KM-Prhsp65-luc plasmid, resulting in being intolerable for transformation.
Author response table 1.
Author response image 3.
Result of luciferase activities before and after transformation of pS0246KM-Prhsp65-luc plasmid. Fifty microliter of cultures were mixed with 50 u L of assay reagents (Luciferase assay system E1500, Promega) and luciferase activity was measured by the luminometer (FilterMax F5, Molecular Devices). Data are shown as mean ± SD of 6-12 colonies
[Reference]
Aoki K. Extracellular mycobacterial DNA-binding protein 1 participates in Mycobacterium-lung epithelial cell interaction through hyaluronic acid. J Biol Chem 279, 39798–39806 (2004).
(5e) Lines 283-186: "To confirm the gene essentiality detected with the HMM analysis, we evaluated the consequent growth inhibition in the knockdown strains of representative universal essential or growth-defect-associated genes, including glcB, inhA, gyrB, and embB.." It is not clear what was the level of suppression of these genes in the respective KD strains. Authors should include the level of suppression of these genes also by qRT-PCR.
Thank you for the comment on the suppression levels of gene expression in knockdown strains of universal essential genes. Following the comment, we have evaluated them by qRT-PCR and we observed comparable levels of knockdown efficiency in the knockdown strains between universally essential genes and strain-specific/accessory essential genes (new Supplementary Fig. 9). Overall, the gene expression was suppressed to 20 - 70% in the knockdown strains compared to the vector control strains that do not express sgRNA.
We have added the data of qRT-PCR of knockdown strains of universal essential genes such as glcB, inhA, gyrB, and embB (new Supplementary Fig. 9). We have revised the Result and Discussion in the manuscript (page 21, lines 367-376; page28, lines 490-497).
(5f) Lines 293-: I am unable to establish any correlation between the growth of the knockdown with Tn insertion reads in the respective genes. For instance, pckA exhibits reduced Tn insertion reads in almost all the strains except in M.i.27, but the effect of its KD on growth is seen only in M.i.198 and M003; glpX exhibits reduced Tn insertion reads in M003, M019, M021 but the effect of its KD on growth is seen only in M003; csd exhibits reduced Tn insertion reads in M.i.198, M003, M019 but the effect of its KD on growth is seen only in M.i.198 and M003. The authors argue that these contradictory phenotypes are due to difficulties in the effective operation of genetically modified systems using foreign genes from different bacterial species in MAC-PD strains (Lines 310-312) or the desired effect on growth could not be observed due to the inability of CRISPRi to yield >99% suppression (Line 314) are not the valid justifications. Indeed, a close look at the RT-PCR data (Figure S5) reveals that pckA levels are ~0.22, 0.5, 0.2, 0.22, 0.2, 0.5, and 0.3 fold relative to sigA in M.i.198, M.i.27, ATCC13950, M018, M019, M003 and M021, respectively, but the effect of its suppression on growth by CRISPRi is seen only in M.i.198 and M003. Secondly, >99% suppression is not a universal prerequisite for all the genes to show growth defect (as might be the case with glcB, inhA, gyrB, and embB genes in this study). Hence, it remains unclear why contrasting results are obtained for most of the genes by TnSeq and CRISPRi.
Thank you for the comments on the issue of inconsistent results between TnSeq and CRISPR-i based knockdown. We acknowledge that some inconsistencies were observed, particularly among strain-dependent/accessory essential or growth-defect associated genes. By contrast, we found consistent data between TnSeq and CRISPR-i based knockdown results of universal essential genes. By obtaining the data of suppression levels of gene expression in the knockdown strains of universal essential genes, we have acknowledged that the low efficiency of knockdown does not explain the reason of the discrepancy between TnSeq and CRISPR-i results because the levels of knockdown efficiency were comparable between strain-dependent/accessory essential genes and universally essential genes.
Although the mechanism has not been fully proven yet only from the current study, we consider that such inconsistent phenotypes with TnSeq and CRISPR-i based knockdown may be related to the recently revealed the bypass mechanism of gene essentiality which is characteristically observed in strain-dependent/accessory essential or growth-defect-associated genes. According to the publication by Rosconi (Nat Microbiol. 2022: new Ref#14) reporting the ‘forced-evolution experiments’ of 36 clinical Streptococcus pneumoniae strains, gene essentiality can be bypassed by several mechanisms including the composition of the accessory genome and pathway rewiring. They recovered successfully knockout mutants from transformation experiments in strain-specific/accessory essential genes such as cytidine monophosphate kinase, a folate pathway enzyme formate tetrahydrofolate ligase and an undecaprenyl phosphate-biosynthesis pathway enzyme farnesyl-diphosphate synthase. The bypassing of gene essentiality could be suggested by observing suppressor mutations and synthetic lethality in knockout strains. By contrast, universal essential genes were reported to fulfill the three categories including high levels of conservation within and often across species, limited genetic diversity, and high and stable expression levels. Consequently, universal essential genes are estimated to be rigid, largely immutable key components to an organism’s survival.
We consider that this is the case with our study on NTM because NTM is pangenomic. The knockdown of universal essential genes resulted in the clear growth suppression; however, the knockdown of strain-dependent/accessory essential genes did not show the consistent growth suppression. We consider that the bypass mechanism of gene essentiality can explain the inconsistent effect of gene silencing of strain-dependent/accessory genes on bacterial growth suppression.
We have added the above-mentioned description in the Discussion (pages 28-29, lines 497-519).
[Reference]
Rosconi, F. et al. A bacterial pan-genome makes gene essentiality strain-dependent and evolvable. Nat Microbiol 7, 1580–1592 (2022).
Minor Comments:
(1) The authors should mention the cut-off of fold-change for all the experiments in the methods section.
Thank you for the comment on the cut-off of fold-change. We set the cut-off of fold-change as adjusted P-value < 0.05. We added the description in the Methods section. (page 41, lines 724-725)
(2) Figure 7 legend (Lines 888-889): "Data are shown as the means {plus minus} SD of triplicate experiments. Data from one experiment representative of three independent experiments (N = 3) are shown."
Figure S3 legend: Data on the growth curves are the means of triplicate experiments. Data from one experiment representative of three independent experiments (N = 3) are shown.
Figure S4 legend: Data are shown as the means {plus minus} SD of triplicate experiments. Data from one experiment representative of two independent experiments (N = 2) are shown.
Figure S5 legend: Gene expression data are the means {plus minus} SD of triplicate experiments. Data from one experiment representative of two independent experiments (N = 2) are shown.
These statements need clarification. Whether multiple independent experiments (biological repeats), each with 2-3 technical replicates performed and the data shown represent one of the multiple biological repeats?
Thank you for the comments on the number of experiments performed and the number of replicates. We have performed two or three independent experiments with 2-3 technical replicates. The data shown represent one of the independent experiments.
(3) Figure 7b: Statistics are missing in the bar graph for growth rate under aerobic conditions.
Thank you for the comment on the statistics of the data regarding growth rate under aerobic conditions. We have added the statistics in the new Fig. 7c.
(4) The authors should check the y-axis in Figure 7b, as it is not clear whether bacteria indeed show a growth rate of 1-3 CFUs/day.
Thank you for the comment on the y-axis in Figure 7b. We have corrected the label of y-axis as “log10[CFUs]/day” in the new Fig. 7c. Additionally, we have corrected the label of y-axis in new Fig. 7a and added the description as “Data are represented as CFUs in 4 μl sample at each timepoint.” in the Fig. 7a legend.
Reviewer #3 (Recommendations For The Authors):
(1) It's notable that strains M001 and MOTT64 failed to undergo a transformation, while seven other strains did. Given that M001, MOTT64, and M019 belong to the same phylogenetic clade, it raises questions about why particular strains within this clade showed different transformation outcomes. It might be valuable for them to discuss this discrepancy in their study.
Thank you for the comment on the difference in capacity of transformation between strains belonging to the same genomic subgroup. Although the direct mechanism determining the competency for foreign DNA has not been elucidated in M. intracellulare and other pathogenic NTM species, several studies on general bacteria suggest the difficulties of introducing foreign DNA into clinical strains compared to the laboratory strains. As suggested in Staphylococcus aureus (Covaglia AR. PNAS. 2010; new ref#55), some clinical strains develop elimination system of foreign nucleic acids such as a type III-like restriction restriction endonuclease. As suggested in gran-negative bacteria (Qin J. Sci Rep. 2022; new Ref#56), there may be some difference in cell surface structures between strains, resulting in the necessity of polymyxin B nonapeptide targeting cell membrane for transforming clinical strains. The efficiency of eliminating foreign DNA may be attributed to various kinds of strain-specific factors including restriction endonuclease, natural CRISPR-interference system and cell wall structures rather than a simple genotypic factor.
We have added the description on the difference of capability in transformation in the Discussion. (page 31, lines 546-558)
[References]
Corvaglia, A.R., François, P., Hernandez, D., Perron, K., Linder, P. & Schrenzel, J. A type III-like restriction endonuclease functions as a major barrier to horizontal gene transfer in clinical Staphylococcus aureus strains. Proc Natl Acad Sci U S A 107, 11954-11958 (2010).
Qin, J., Hong, Y., Pullela, K., Morona, R., Henderson, I.R. & Totsika, M. A method for increasing electroporation competence of Gram-negative clinical isolates by polymyxin B nonapeptide. Sci Rep 12,:11629 (2022).
(2) The authors should consider specifying M. intracellulare in their title.
Thank you for the comment on the manuscript title. Following the comments from all Reviewers, we have modified the title as “Functional genomics reveals strain-specific genetic requirements conferring hypoxic growth in Mycobacterium intracellulare”.
-

-

eLife assessment
This descriptive study reports the genetic requirements for growth and fitness of multiple clinical strains of a relatively understudied species of mycobacteria, Mycobacterium intracellulare. The findings are valuable however, the study is incomplete as the primary claims related to hypoxia adaptation need additional experimental support and data presentation requires more clarity. The work will be of interest to microbiologists.
-

Reviewer #1 (Public Review):
Summary:
Tateishi et al. report a Tn-seq-based analysis of genetic requirements for growth and fitness in 8 clinical strains of Mycobacterium intracellulare Mi), and compare the findings with a type strain ATCC13950. The study finds a core set of 131 genes that are essential in all nine strains, and therefore are reasonably argued as potential drug targets. Multiple other genes required for fitness in clinical isolates have been found to be important for hypoxic growth in the type strain.
Strengths:
The study has generated a large volume of Tn-seq datasets of multiple clinical strains of Mi from multiple growth conditions, including from mouse lungs. The dataset can serve as an important resource for future studies on Mi, which despite being clinically significant remains a relatively understudied species of …
Reviewer #1 (Public Review):
Summary:
Tateishi et al. report a Tn-seq-based analysis of genetic requirements for growth and fitness in 8 clinical strains of Mycobacterium intracellulare Mi), and compare the findings with a type strain ATCC13950. The study finds a core set of 131 genes that are essential in all nine strains, and therefore are reasonably argued as potential drug targets. Multiple other genes required for fitness in clinical isolates have been found to be important for hypoxic growth in the type strain.
Strengths:
The study has generated a large volume of Tn-seq datasets of multiple clinical strains of Mi from multiple growth conditions, including from mouse lungs. The dataset can serve as an important resource for future studies on Mi, which despite being clinically significant remains a relatively understudied species of mycobacteria.
Weaknesses:
The paper lacks clarity in data presentation and organization. For example, some of the key data on cfu counts of clinical Mi strains in a mouse model can be presented along with the Tn-seq dataset in Figure 6, the visualization of which can be improved with volcano plots. etc. Improvement in data visualization is perhaps necessary throughout the paper.
The primary claim of the study that the clinical strains are better adapted for hypoxic growth is not well-supported by the data presented in Figure 7.
The title of the paper is misleading as the study doesn't provide any mechanistic aspect of hypoxic adaptation in Mi.
-

Reviewer #2 (Public Review):
Summary:
In the study titled "Functional genomics reveals the mechanism of hypoxic adaptation in nontuberculous mycobacteria" by Tateishi et al., the authors have used TnSeq to identify the common essential and growth-defect-associated genes that represent the genomic diversity of clinical M. intracellulare strains in comparison to the reference type strain. By estimating the frequency of Tn insertion, the authors speculate that genes involved in gluconeogenesis, the type VII secretion system, and cysteine desulfurase are relatively critical in the clinical MAC-PD strains than in the type strain, both for the extracellular survival and in a mouse lung infection model.
Based on their analysis, the authors proposed to identify the mechanism of hypoxic adaptation in nontuberculous mycobacteria (NTM) which offer …
Reviewer #2 (Public Review):
Summary:
In the study titled "Functional genomics reveals the mechanism of hypoxic adaptation in nontuberculous mycobacteria" by Tateishi et al., the authors have used TnSeq to identify the common essential and growth-defect-associated genes that represent the genomic diversity of clinical M. intracellulare strains in comparison to the reference type strain. By estimating the frequency of Tn insertion, the authors speculate that genes involved in gluconeogenesis, the type VII secretion system, and cysteine desulfurase are relatively critical in the clinical MAC-PD strains than in the type strain, both for the extracellular survival and in a mouse lung infection model.
Based on their analysis, the authors proposed to identify the mechanism of hypoxic adaptation in nontuberculous mycobacteria (NTM) which offer promising drug targets in the strains causing clinical Mycobacterium avium-intracellulare complex pulmonary disease (MAC-PD).
Strengths:
A major strength of the manuscript is the performance of the exhaustive set of TnSeq experiments with multiple strains of M. intracellulare during in vitro growth and animal infection.
Weaknesses:
(1) The study suffers from the authors' preconceived bias toward a small subset of genes involved in hypoxic pellicle formation in ATCC13950.
(2) An important set of data with the ATCC13950 reference strain is missing in the mouse infection study. In the absence of this, it is difficult to establish whether the identified genes are critical for infection/intracellular proliferation, specifically in the clinical isolates that are relatively more adapted for hypoxia.
(3) Statistical enrichment analysis of gene sets by GSEA wrongly involves genes required for hypoxic pellicle formation in ATCC13950 together with the gene sets found essential in the clinical MAC-PD strains, to claim that a significant % of genes belong to hypoxia-adaptation pathways. It could be factually incorrect because a majority of these might overlap with those found critical for the in vitro survival of MAC-PD strains (and may not be related to hypoxia).
(4) Validation of mouse infection experiments with individual mutants is missing.
(5) Phenotypes with TnSeq and CRISPRi-based KD exhibit poor correlation with misleading justifications by the authors.
In summary, this study is unable to provide mechanistic insights into why and how different MAC-PD mutant strains exhibit differential survival (in vitro and in animals) and adaptation to hypoxia. It remains to understand why the clinical strains show better adaptation to hypoxia and what is the impact of other stresses on their growth rates.
-

Reviewer #3 (Public Review):
Summary:
The study by Tateishi et al. utilized TnSeq in nine genetically diverse M. intracellulare strains, identifying 131 common essential and growth-defect-associated genes across those strains, which could serve as potential drug targets. The authors also provided an overview of the differences in gene essentiality required for hypoxic growth between the reference strain and the clinical strains. Furthermore, they validated the universal and accessory/strain-dependent essential genes by knocking down their expression using CRISPRi technique. Overall, this study offers a comprehensive assessment of gene requirements in different clinical strains of M. intracellular.
(1) The rationale for using ATCC13950 versus clinical strains needs to be clarified. The reference strain ATCC13950 was obtained from the …
Reviewer #3 (Public Review):
Summary:
The study by Tateishi et al. utilized TnSeq in nine genetically diverse M. intracellulare strains, identifying 131 common essential and growth-defect-associated genes across those strains, which could serve as potential drug targets. The authors also provided an overview of the differences in gene essentiality required for hypoxic growth between the reference strain and the clinical strains. Furthermore, they validated the universal and accessory/strain-dependent essential genes by knocking down their expression using CRISPRi technique. Overall, this study offers a comprehensive assessment of gene requirements in different clinical strains of M. intracellular.
(1) The rationale for using ATCC13950 versus clinical strains needs to be clarified. The reference strain ATCC13950 was obtained from the abdominal lymph node of a patient around 10 years ago and is therefore considered a clinical strain that has undergone passages in vitro. How many mutations have accumulated during these in vitro passages? Are these mutations significant enough to cause the behavior of ATCC13950 to differ from other recently sampled clinical strains? From the phylogenetic tree, ATCC13950 is located between M018 and M.i.27. Did the authors observe a similarity in gene essentiality between ATCC13950 and its neighbor strains? What is the key feature that separates ATCC13950 from these clinical strains? The authors should provide a strong rationale for how to interpret the results of this comparison in a clinical or biological context.
(2) Regarding the 'nine representative strains of M. intracellulare with diverse genotypes in this study,' how were these nine strains selected? To what extent do they represent the genetic diversity of the M. intracellulare population? A phylogenetic tree illustrating the global genetic diversity of the M. intracellulare population, with these strains marked on it, would be important to demonstrate their genetic representativeness.
(3) The authors observed a considerable amount of differential gene requirements in clinical strains. However, the genetic underpinning underlying the differential requirement of genes in clinical strains was not investigated or discussed. Because M. intracellulare has a huge number of accessory genes, the authors should at least check whether the differential requirement could be explained by the existence of a second copy of functional analogous genes or duplications.
(4) Growth in aerobic and hypoxic conditions: The authors concluded that clinical strains are better adapted to hypoxia, as reflected by their earlier entry into the log phase. They presented the 'Time at midpoint' and 'Growth rate at midpoint.' However, after reviewing the growth curves, I noticed that ATCC13950 had a longer lag phase compared to other strains under hypoxic conditions, and its phylogenetic neighbor M018 also had a longer lag phase. Hence, I do not believe a conclusion can be drawn that clinical strains are better adapted to hypoxia, as this behavior could be specific to a particular clade. It's also possible that the ATCC13950 strain has adapted to aerobic growth. I would suggest that the authors include growth curves in the main figures. The difference in 'Time at midpoint' could be attributed to several factors, and visualizing the growth curves would provide additional context and clarity.
(5) Lack of statistical statement: The authors emphasized the role of pellicle-formation-associated genes in strain-dependent essential and accessory essential genes. Additionally, the authors observed that 10% of the genes required for mouse infection are also required for hypoxic pellicle formation. However, these are merely descriptive statements. There is no enrichment analysis to justify whether pellicle-formation-associated genes are significantly enriched in these groups.
-