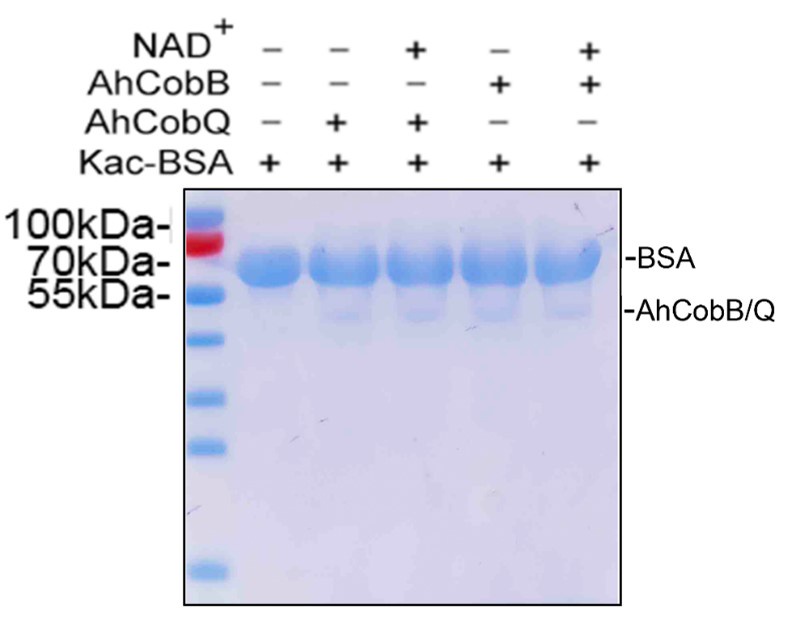
Aeromonas hydrophila CobQ is a new type of NAD+- and Zn2+-independent protein lysine deacetylase
Curation statements for this article:-
Curated by eLife
eLife Assessment
In this valuable study, the authors studied a novel Zn2+- and NAD+-independent KDAC protein, AhCobQ, in Aeromonas hydrophila, which lacks homology with eukaryotic counterparts, thus underscoring its unique evolutionary trajectory within the bacterial domain. They attempt to demonstrate deacetylase activity, however, whilst the revised manuscript has been improved, significant aspects of the data are still incomplete and require further refinement. The work will be of interest to microbiologists studying metabolism and post-translational modifications.
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Protein N Ɛ -lysine acetylation (Kac) modifications play crucial roles in diverse physiological and pathological functions in cells. In prokaryotic cells, there are only two types of lysine deacetylases (KDACs) that are Zn 2+ - or NAD + -dependent. In this study, we reported a protein, AhCobQ, in Aeromonas hydrophila ATCC 7966 that presents NAD + - and Zn 2+ -independent KDAC activity. Furthermore, its KDAC activity is located in an unidentified domain (from 195 to 245 aa). Interestingly, AhCobQ has no homology with current known KDACs, and no homologous protein was found in eukaryotic cells. A protein substrate analysis showed that AhCobQ has specific protein substrates in common with other known KDACs, indicating that these KDACs can dynamically co-regulate the states of Kac proteins. Microbiological methods employed in this study affirmed AhCobQ’s positive regulation of isocitrate dehydrogenase (ICD) enzymatic activity at the K388 site, implicating AhCobQ in the modulation of bacterial enzymatic activities. In summary, our findings present compelling evidence that AhCobQ represents a distinctive type of KDAC with significant roles in bacterial biological functions.
Article activity feed
-

-

eLife Assessment
In this valuable study, the authors studied a novel Zn2+- and NAD+-independent KDAC protein, AhCobQ, in Aeromonas hydrophila, which lacks homology with eukaryotic counterparts, thus underscoring its unique evolutionary trajectory within the bacterial domain. They attempt to demonstrate deacetylase activity, however, whilst the revised manuscript has been improved, significant aspects of the data are still incomplete and require further refinement. The work will be of interest to microbiologists studying metabolism and post-translational modifications.
-

Reviewer #1 (Public review):
Summary:
This study by Wang et al. identifies a new type of deacetylase, CobQ, in Aeromonas hydrophila. Notably, the identification of this deacetylase reveals a lack of homology with eukaryotic counterparts, thus underscoring its unique evolutionary trajectory within the bacterial domain.
Strengths:
The manuscript convincingly illustrates CobQ's deacetylase activity through robust in vitro experiments, establishing its distinctiveness from known prokaryotic deacetylases. Additionally, the authors elucidate CobQ's potential cooperation with other deacetylases in vivo to regulate bacterial cellular processes. Furthermore, the study highlights CobQ's significance in the regulation of acetylation within prokaryotic cells.
Weaknesses:
The problem I raised has been well resolved. I have no further questions.
-

Reviewer #2 (Public review):
In recent years, lots of researchers tried to explore the existence of new acetyltransferase and deacetylase by using specific antibody enrichment technologies and high resolution mass spectrometry. Here is an example for this effort. Yuqian Wang et al. studied a novel Zn2+- and NAD+-independent KDAC protein, AhCobQ, in Aeromonas hydrophila. They studied the biological function of AhCobQ by using biochemistry method and MS identification technology to confirm it. These results extended our understanding of the regulatory mechanism of bacterial lysine acetylation modifications. However, I find this conclusion is a little speculative, and unfortunately it also doesn't totally support the conclusion as the authors provided.
Major concerns:
-It is a little arbitrary to come to the title "Aeromonas hydrophila …
Reviewer #2 (Public review):
In recent years, lots of researchers tried to explore the existence of new acetyltransferase and deacetylase by using specific antibody enrichment technologies and high resolution mass spectrometry. Here is an example for this effort. Yuqian Wang et al. studied a novel Zn2+- and NAD+-independent KDAC protein, AhCobQ, in Aeromonas hydrophila. They studied the biological function of AhCobQ by using biochemistry method and MS identification technology to confirm it. These results extended our understanding of the regulatory mechanism of bacterial lysine acetylation modifications. However, I find this conclusion is a little speculative, and unfortunately it also doesn't totally support the conclusion as the authors provided.
Major concerns:
-It is a little arbitrary to come to the title "Aeromonas hydrophila CobQ is a new type of NAD+- and Zn2+-independent protein lysine deacetylase in prokaryotes." It should be modified to delete the "in the prokaryotes" except that the authors get new more evidence in the other prokaryotes for the existence of the AhCobQ.
-I was confused about the arrangement of the supplementary results. Because there are no citations for Figures S9-S19.
-Same to the above, there are no data about Tables S1-S6.
-All the load control is not integrated. Please provide all of the load controls with whole PAGE gel or whole membrane western blot results. Without these whole results, it is not convincing to come the conclusion as the authors mentioned in the context.
-Thoroughly review the materials & methods section. It is unclear to me what exactly the authors describe in the method. All the experimental designs and protocols should be described in detail, including growth conditions, assay conditions, and purification conditions, etc.
-Include relevant information about the experiments performed in the figure legends, such as experimental conditions, replicates, etc. Often it is not clear what was done based on the figure legend description. -

Author response:
The following is the authors’ response to the current reviews.
Public Reviews:
Reviewer #1 (Public review):
Summary:
This study by Wang et al. identifies a new type of deacetylase, CobQ, in Aeromonas hydrophila. Notably, the identification of this deacetylase reveals a lack of homology with eukaryotic counterparts, thus underscoring its unique evolutionary trajectory within the bacterial domain.
Strengths:
The manuscript convincingly illustrates CobQ's deacetylase activity through robust in vitro experiments, establishing its distinctiveness from known prokaryotic deacetylases. Additionally, the authors elucidate CobQ's potential cooperation with other deacetylases in vivo to regulate bacterial cellular processes. Furthermore, the study highlights CobQ's significance in the regulation of acetylation within prokaryotic …
Author response:
The following is the authors’ response to the current reviews.
Public Reviews:
Reviewer #1 (Public review):
Summary:
This study by Wang et al. identifies a new type of deacetylase, CobQ, in Aeromonas hydrophila. Notably, the identification of this deacetylase reveals a lack of homology with eukaryotic counterparts, thus underscoring its unique evolutionary trajectory within the bacterial domain.
Strengths:
The manuscript convincingly illustrates CobQ's deacetylase activity through robust in vitro experiments, establishing its distinctiveness from known prokaryotic deacetylases. Additionally, the authors elucidate CobQ's potential cooperation with other deacetylases in vivo to regulate bacterial cellular processes. Furthermore, the study highlights CobQ's significance in the regulation of acetylation within prokaryotic cells.
Weaknesses:
The problem I raised has been well resolved. I have no further questions.
Thanks for your valuable comments very much.
Reviewer #2 (Public review):
In recent years, lots of researchers tried to explore the existence of new acetyltransferase and deacetylase by using specific antibody enrichment technologies and high resolution mass spectrometry. Here is an example for this effort. Yuqian Wang et al. studied a novel Zn2+- and NAD+-independent KDAC protein, AhCobQ, in Aeromonas hydrophila. They studied the biological function of AhCobQ by using biochemistry method and MS identification technology to confirm it. These results extended our understanding of the regulatory mechanism of bacterial lysine acetylation modifications. However, I find this conclusion is a little speculative, and unfortunately it also doesn't totally support the conclusion as the authors provided.
Major concerns:
- It is a little arbitrary to come to the title "Aeromonas hydrophila CobQ is a new type of NAD+- and Zn2+-independent protein lysine deacetylase in prokaryotes." It should be modified to delete the "in the prokaryotes" except that the authors get new more evidence in the other prokaryotes for the existence of the AhCobQ.
Thank you for your suggestion. However, I believe there has been some confusion regarding the title. In the revised manuscript we have already updated the title to: "Aeromonas hydrophila CobQ is a new type of NAD+- and Zn2+-independent protein lysine deacetylase."
This title does not include the phrase "in prokaryotes," as you mentioned. We kindly suggest verifying the version of the manuscript that was reviewed to ensure you are reviewing the most recent changes.
- I was confused about the arrangement of the supplementary results. Because there are no citations for Figures S9-S19.
Thank you for your feedback. It appears there may have been a misunderstanding, possibly due to reviewing an outdated version of the manuscript. In the revised manuscript we revised the supplementary figures and now have only 12 figures, all of which are correctly cited in the manuscript on pages 12 to 15. Below is a detailed list of the updated figure citations:
Figures S1: page 8, line 148;
Figures S2: page 9, line 168;
Figures S3 and S4: page 10, line 178;
Figures S5: page 10, line 186;
Figures S6: page 10, line 189;
Figures S7: page 12, line 221;
Figures S8-S10: page 13, line 245;
Figures S11: page 11, line 282;
Figures S12: page 15, line 286
- Same to the above, there are no data about Tables S1-S6.
Thank you for your attention to the supplementary materials. As with the figures, we have already uploaded the data for Tables S1-S6 in the revised manuscript on November 19, 2024, and properly cited Tables S1 – S6 in the manuscript. Below is the citation information:
Tables S1: page 10, line 194;
Tables S2: page 13, line 245;
Tables S3: page 21, line 438;
Tables S4: page 22, line 439;
Tables S5: page 22, line 445;
Tables S6: page 27, line 564.
Please note that Tables S3 – S4 include the chemical reagents, primers, and other experimental materials, which are not intended to be cited in the results section.)
- All the load control is not integrated. Please provide all of the load controls with whole PAGE gel or whole membrane western blot results. Without these whole results, it is not convincing to come the conclusion as the authors mentioned in the context.
Thank you for your comment. Please note that the full membrane western blot results were included in the revised manuscript. We hope this satisfies your request. If you need further clarification or additional data, please do not hesitate to let us know.
- Thoroughly review the materials & methods section. It is unclear to me what exactly the authors describe in the method. All the experimental designs and protocols should be described in detail, including growth conditions, assay conditions, and purification conditions, etc.
Thank you for your valuable suggestion. In response to your comment and previous feedback, we have alredy revised the Materials & Methods section thoroughly in the revised manuscript. The experimental details, including growth conditions, assay protocols, and purification procedures, are described in full on pages 22 to 30 of the revised manuscript.
- Include relevant information about the experiments performed in the figure legends, such as experimental conditions, replicates, etc. Often it is not clear what was done based on the figure legend description.
Thank you very much for your detailed feedback and suggestions. We have made sure to describe what each data point represents in the figure legends, as per the previous feedback. However, we would like to clarify that while we have provided detailed descriptions in the legends, the inclusion of every specific experimental condition in the figure legends could result in redundancy, as these details are already thoroughly outlined in the Materials & Methods section.
We hope this explanation addresses your concern.
Recommendations for the authors:
Reviewer #1 (Recommendations for the authors):
I have no further revision comments.
Thank you very much.
Reviewer #2 (Recommendations for the authors):
I carefully read the point-to-point response from the author. Although they listed lots of the reasons for the ugly results, it still can not persuade me to accept their conclusions. While, as I know, it is impossible to reject their work in eLife as it was sent out for peer-review. I also can't accuse them of being wrong, but I have my opinion on this point. That is not the results, but the attitude.
Thank you for your feedback. However, I must express some concerns regarding the nature of your comments. Based on the issues you've raised, it seems that you may have reviewed an outdated version of the manuscript. In the updated revision we addressed all the points you've raised, including the figure and table citations, experimental methods, and data integration.
We understand that differing opinions are part of the peer-review process, but we respectfully believe that your conclusion regarding our attitude is based on a misunderstanding, possibly caused by reviewing an incorrect version of the manuscript. We have always strived to approach this manuscript with utmost professionalism and have diligently responded to each of your concerns.
We sincerely suggest reviewing the latest version of our manuscript, and we welcome any further constructive feedback. We hope this clarifies any misunderstandings and look forward to your continued support.
Thank you for your time and thoughtful consideration.
The following is the authors’ response to the original reviews.
Public Reviews:
Reviewer #1 (Public review):
Summary:
This study by Wang et al. identifies a new type of deacetylase, CobQ, in Aeromonas hydrophila. Notably, the identification of this deacetylase reveals a lack of homology with eukaryotic counterparts, thus underscoring its unique evolutionary trajectory within the bacterial domain.
Strengths:
The manuscript convincingly illustrates CobQ's deacetylase activity through robust in vitro experiments, establishing its distinctiveness from known prokaryotic deacetylases. Additionally, the authors elucidate CobQ's potential cooperation with other deacetylases in vivo to regulate bacterial cellular processes. Furthermore, the study highlights CobQ's significance in the regulation of acetylation within prokaryotic cells.
Weaknesses:
The problem I raised has been well resolved. I have no further questions.
Reviewer #2 (Public review):
In recent years, lots of researchers tried to explore the existence of new acetyltransferase and deacetylase by using specific antibody enrichment technologies and high resolution mass spectrometry. Here is an example for this effort. Yuqian Wang et al. studied a novel Zn2+- and NAD+-independent KDAC protein, AhCobQ, in Aeromonas hydrophila. They studied the biological function of AhCobQ by using biochemistry method and MS identification technology to confirm it. These results extended our understanding of the regulatory mechanism of bacterial lysine acetylation modifications. However, I find this conclusion is a little speculative, and unfortunately, it also doesn't totally support the conclusion as the authors provided.
Reviewer #3 (Public review):
Summary:
This study reports on a novel NAD+ and Zn2+-independent protein lysine deacetylase (KDAC) in Aeromonas hydrophila, termed as AhCobQ (AHA_1389). This protein is annotated as a CobQ/CobB/MinD/ParA family protein and does not show similarity with known NAD+-dependent or Zn2+-dependent KDACs. The authors showed that AhCobQ has NAD+ and Zn2+-independent deacetylase activity with acetylated BSA by western blot and MS analyses. They also provided evidence that the 195-245 aa region of AhCobQ is responsible for the deacetylase activity, which is conserved in some marine prokaryotes and has no similarity with eukaryotic proteins. They identified target proteins of AhCobQ deacetylase by proteomic analysis and verified the deacetylase activity using site-specific Kac proteins. Finally, they showed that AhCobQ activates isocitrate dehydrogenase by deacetylation at K388.
Strengths:
The finding of a new type of KDAC has a valuable impact on the field of protein acetylation. The characters (NAD+ and Zn2+-independent deacetylase activity in an unknown domain) shown in this study are very unexpected.
Weaknesses:
(1) The characters (NAD+ and Zn2+-independent deacetylase activity in an unknown domain) shown in this study are very unexpected. To convince readers, MSMS data must be necessary to accurately detect (de)acetylation at the target site in the deacetylase activity assay. The authors showed the MSMS data in assays with acetylated BSA, but other assays only rely on western blot.
(2) They prepared site-specific Kac proteins and used them in deacetylase activity assays. Incorporation of acetyllysine at the target site should be confirmed by MSMS and shown as supplementary data.
(3) The authors imply that the 195-245 aa region of AhCobQ may represent a new domain responsible for deacetylase activity. The feature of the region would be of interest but is not sufficiently described in Figure 5. The amino acid sequence alignments with representative proteins with conserved residues would be informative. It would be also informative if the modeled structure predicted by AlphaFold is shown and the structural similarity with known deacetylases is discussed.
Recommendations for the authors:
Reviewer #1 (Recommendations for the authors):
The problem I raised has been well resolved. I have no further questions.
Reviewer #2 (Recommendations for the authors):
Questions to response of"-The load control is not all integrated. All of the load controls with whole PAGE gel or whole membrane western blot results should be provided. Without these whole results, it is not convincing to come to the conclusion that the authors have."
Just as the Authors answered. The Coomassie Blue R-350 staining outcomes from the PVDF membranes. That is a good control for the experiment. However, I still have several questions about it:
(1) The first is the quality of these Western blot. Why all the bands of these Western blot is so ugly? To tell the truth, it is very difficult to come to a conclusion from these poor western blots.
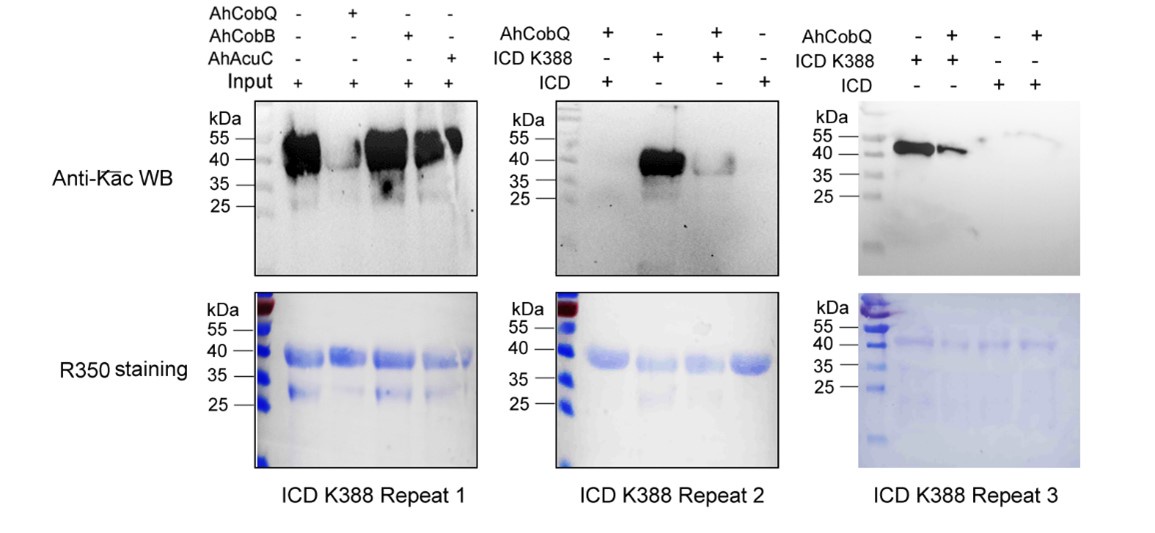
We appreciate your feedback regarding the quality of the Western blots presented in Figure 7. We believe the “ugly bands” you referred to reflect our results validating the functions of CobQ through the use of recombinant site-specific Kac protein substrates.
In our study, we meticulously engineered these recombinant site-specific Kac proteins using a two-plasmid system, based on foundational research published in Nature Chemical Biology (2017, 13(12): 1253-1260), which introduced the genetic encoding of Nε-acetyllysine into recombinant proteins. However, we faced a common challenge: protein truncation due to premature translation termination at the reassigned codon. This issue not only hampers protein yields, as discussed in ChemBioChem (2017, 18(20): 1973-1983), but also contributes to the suboptimal appearance of the Western blot results.
Despite conducting at least two independent repetitions for the Western blot analysis of the site-specific Kac proteins, which yielded consistent results, we recognize that the overall quality remains less than ideal. This variability is inherently related to the characteristics of the target proteins. Nevertheless, the primary aim of our manuscript is to validate the novel deacetylase activity of CobQ. We have provided multiple lines of evidence, including mass spectrometry (MS/MS) and Western blot analyses, to substantiate this claim. In response to your comments, we have decided to remove the ambiguous Western blot results from Figure 7, retaining only four figures that demonstrate significant differences across at least two independent replicates (Author response images 1-5). Additionally, we have included four biological replicates of the Western blot results for ICD Kac388 + CobQ in the supplementary materials (Author response image 5) to further validate the deacetylase function of CobQ.
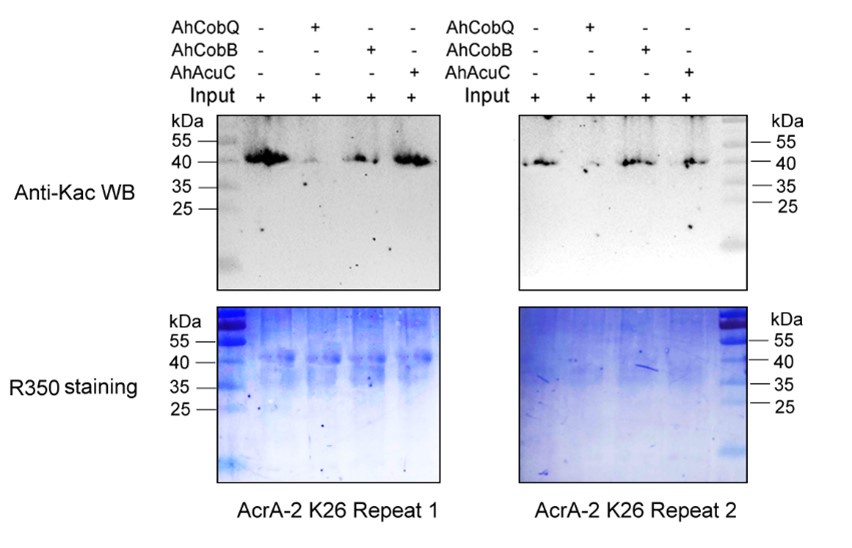
Author response image 1.
Western blot validation of the Kac26 AcrA-2 protein substrates regulated by the three KDACs in two biological replicates.
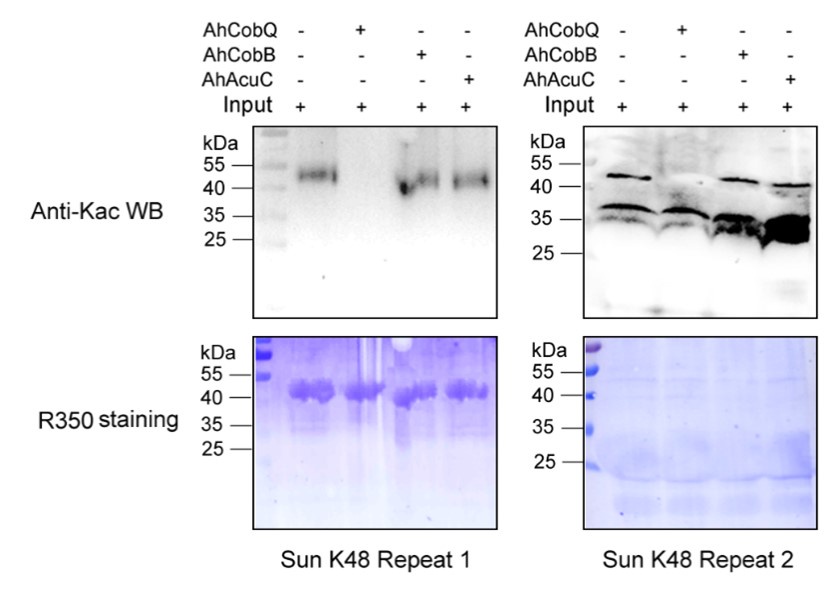
Author response image 2.
Western blot validation of the Kac48 Sun protein substrates regulated by the three KDACs in two biological replicates.
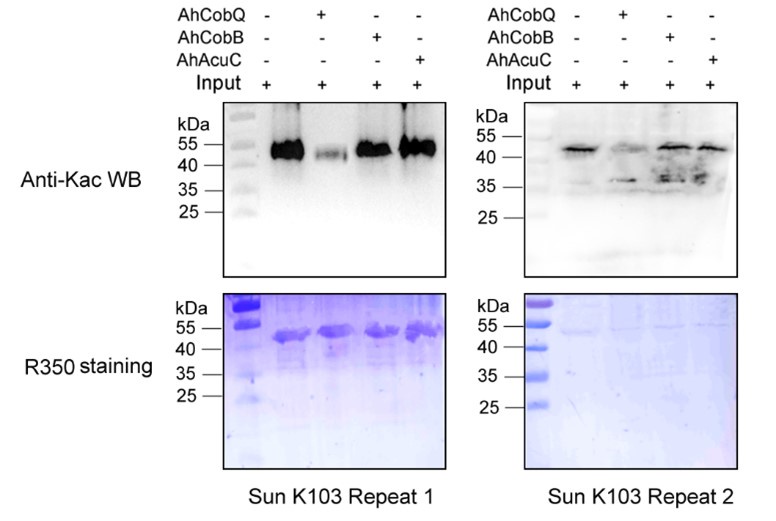
Author response image 3.
Western blot validation of the Kac103 Sun protein substrates regulated by the three KDACs in two biological replicates.
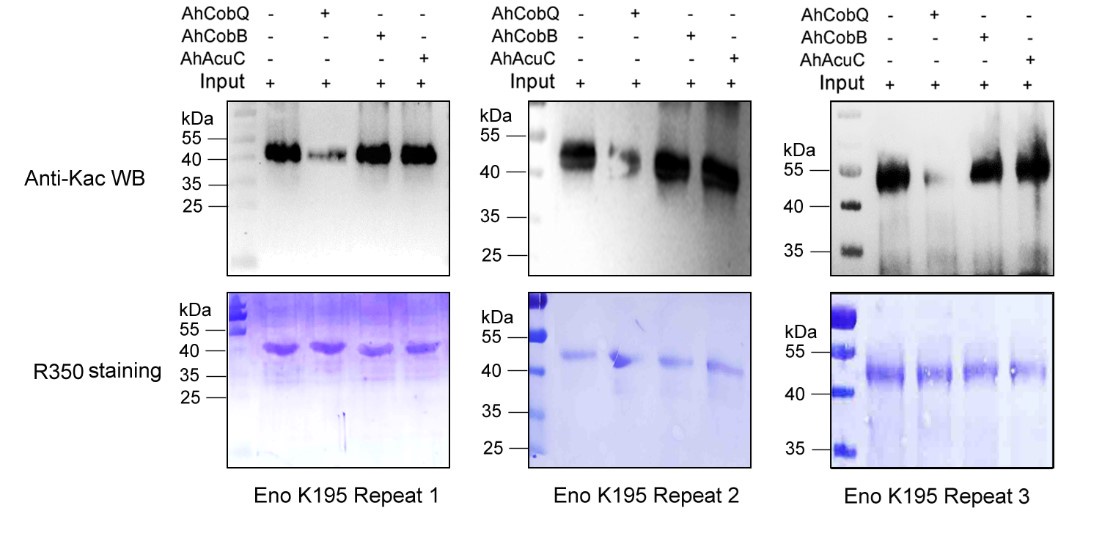
Author response image 4.
Western blot validation of the Kac195 Eno protein substrates regulated by the three KDACs in three biological replicates.
Author response image 5.
Western blot validation of the Kac388 ICD protein substrates regulated by AhCobQ in this study. Each sample was independently repeated at least three time.
(2) The second is why some of the results are not from the same PVDF by comparing the Coommassie staining with the WB results just as authors responded. For example, the HrpA-K816 (ac), Eno-K195 (ac), ArcA-2-K26 (ac), ArcA-2-K26 (ac), IscS-K93(ac), A0KJ75-K81(ac), GyrB-K331(ac), GyrB-K449(ac), FtsA-K320(ac), FtsA-K409(ac), RecA-K279(ac), and the RecA-K306(ac). All of them are clearly not from the same staining results of PVDF membrane but from a new PVDF membrane.
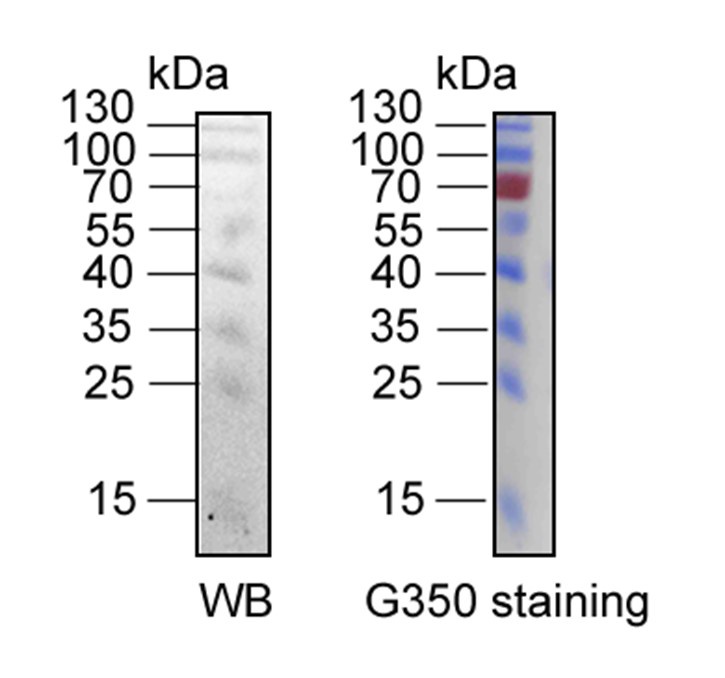
We assure you that the R-350 stained PVDF membranes originate from the same Western blot membranes. However, we acknowledge that visual discrepancies may arise due to differences in imaging techniques. The Western blot results were scanned using a ChemiDoc MP (Bio-Rad, Hercules, CA, USA), while the Coomassie R-350 stained PVDF membranes were captured using a standard camera. These differences can create a misleading appearance, making it seem as though they come from different membranes.
It is also important to note that the intensity of the protein marker cannot be directly compared between the two imaging methods. As illustrated in Author response image 6, the protein marker at 70 kDa is clearly detectable in the Coomassie R-350 image, whereas it may not be as apparent in the Western blot result due to inherent differences in detection sensitivity.
Author response image 6.
The comparison of Western blotting and R-350 strained results of same protein marker in the same PVDF membrane. The protein marker located at 70 kDa can be detected easily in Coomassie R-350, while is difficult to display in WB result.
Additionally, we have removed some of the so-called "ugly" Western blot results in the updated manuscript and provided the original full film of the relevant images as an attachment. This documentation demonstrates that all the data you referenced originate from the same film, as shown in Figures 1-5.
(3) The third is why there is no replication for all these WB results. We should draw a conclusion with serious attitude, but not from the only one repeat, even say nothing about the poor results.
Thank you for your valuable suggestion. In the second version of the manuscript, we have included the original full film of the relevant images. While we previously explained the reasons behind the "ugly" Western blot results, we have decided to remove some, or even all, of these results from Figure 7 in the updated version. The related images will be updated in the supplementary materials (Figures 1-5 in responding letter and Figure 7 in the revised manuscript).
Furthermore, we have provided a more detailed discussion regarding the poor results in the updated manuscript to ensure clarity and transparency. We appreciate your understanding and hope these changes meet your expectations.
Questions to response of " L174-187, L795 (Please show the whole membrane (or PAGE gel) of the loading control of CobB, and CobQ, except for the Kac-BSA)".
(1) As we all educated that there is no control, and no biology. Where is the band of CobQ? Why do not stain the same PVDF membrane with R-350 staining but with a new membrane?
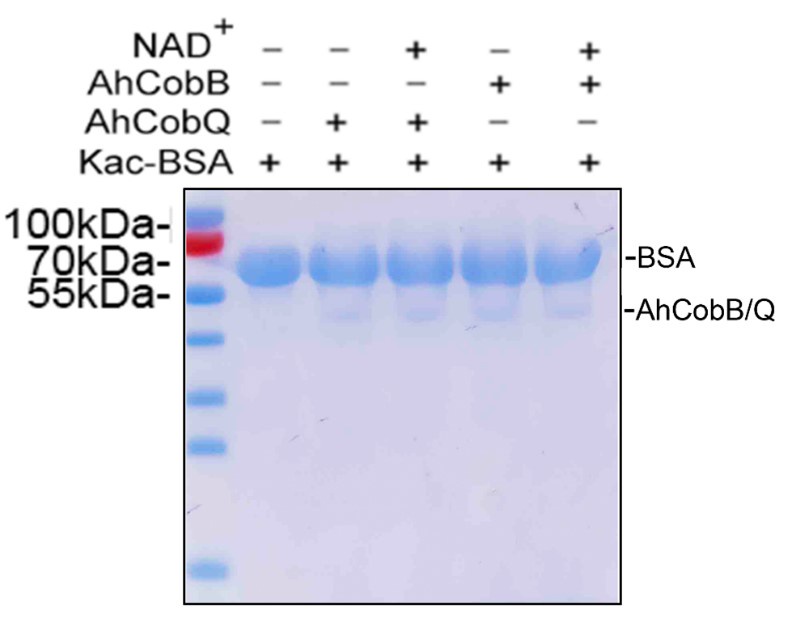
Thank you for your insightful feedback. As noted in our previous response, the absence of visible bands for AhCobQ and AhCobB on the Coomassie R-350 stained PVDF membrane is primarily due to the low loading amounts and protein loss during the Western blotting process.
To reinforce our findings, we repeated the analysis of the protein samples via SDS-PAGE, using the same loading quantity as in the previous Western blot shown in Figure 2 of the manuscript. As illustrated in Author response image 7, the bands for CobB and CobQ are discernible, albeit with significantly lower intensities compared to the Kac-BSA bands. Upon examining the full Coomassie R-350 stained PVDF membranes provided in Supplementary Material 1, we observe that the CobB and CobQ bands are not easily visible. This aligns with your observations and can be attributed to potential protein loss during the transfer from SDS-PAGE to the PVDF membrane.
Author response image 7.
The SDS-PAGE gel displayed the loading amounts of Kac-BSA and CobB/CobQ.
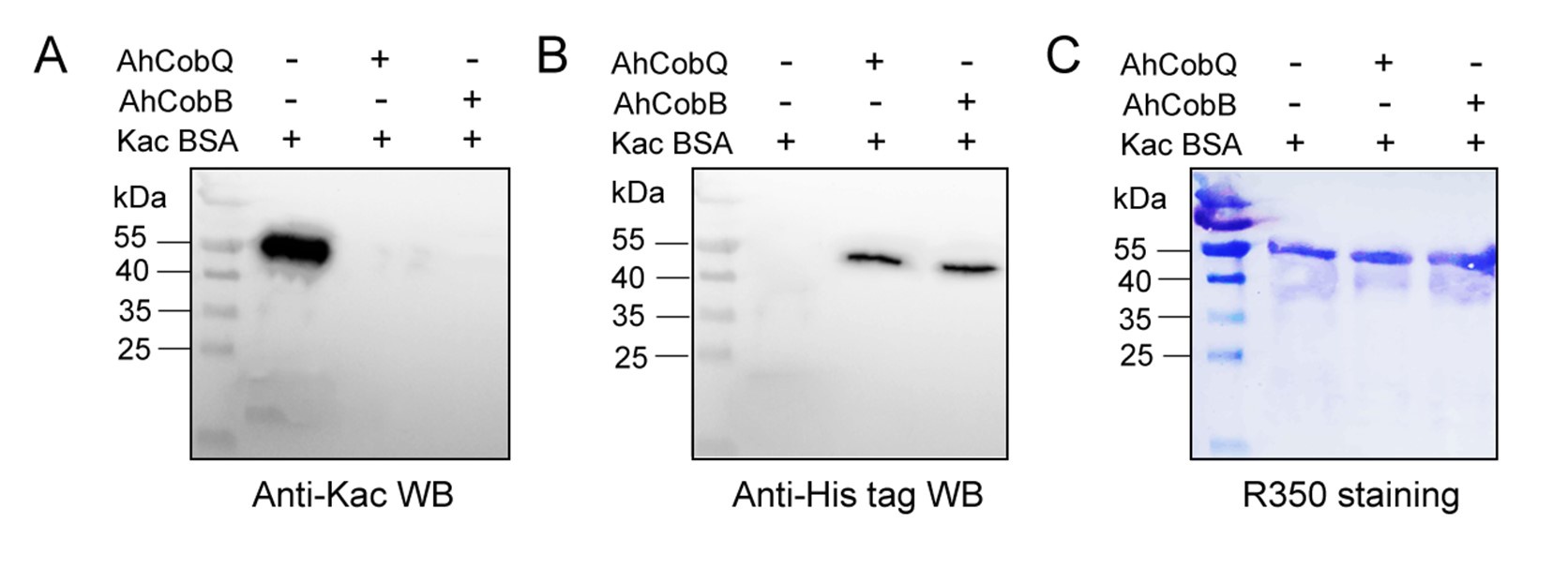
To enhance the visibility of the CobQ/CobB bands, we increased the loading of CobQ/CobB in a new Western blot experiment, using 2 µg of Kac-BSA in combination with 0.8 µg of CobQ/CobB. As shown in Figure 8, while the increasing amounts of Kac-BSA resulted in a more blurred signal, the bands for the recombinant CobQ and CobB proteins were clearly detectable. This indicates that both proteins were indeed involved in the in vitro protein deacetylation assay.
Author response image 8.
Western blot verified the deacetylase activity assay of AhCobQ and AhCobB on Kac-BSA.
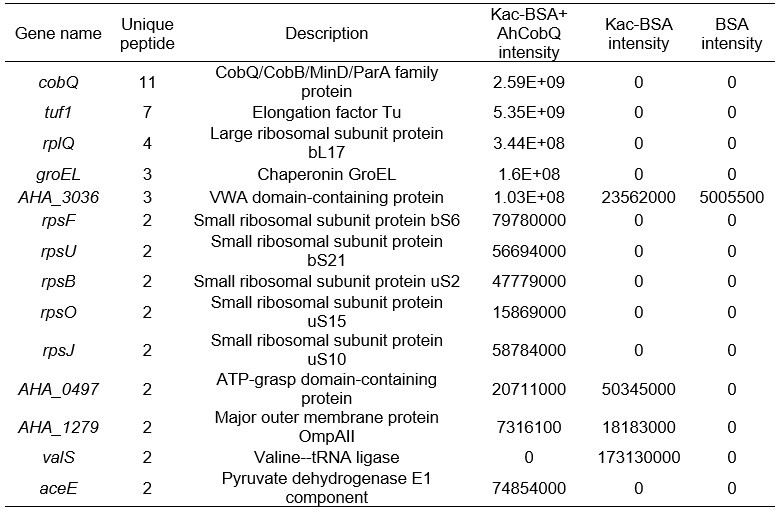
Furthermore, we conducted a mass spectrometry analysis comparing Kac-BSA and Kac-BSA incubated with CobQ, as well as BSA without acetylation, against the A. hydrophila database with a cut-off of unique matched peptides >1. It is challenging to completely avoid contaminant detection during protein purification, especially when using high-resolution mass spectrometry. Our findings revealed that CobQ has the highest number of unique matched peptides (Author response table 1), while contaminants such as AHA_3036, AHA_0497, AHA_1279, and valS could be excluded, as they were present in Kac-BSA or BSA samples. Additionally, Tuf1, RplQ, GroEL, RpsF, RpsU, RpsB, RpsO, and RpsJ are known ribosomal subunits or chaperonins that are abundantly expressed in cells and may interact with various proteins, leading to contaminant detection.
Author response table 1.
LC MS/MS results of selected peptide quantification among Kac-BSA and Kac-BSA incubated with CobQ and BSA without acetylation against A. hydrophila database (unique matched peptides>1).
Although AceE, a pyruvate dehydrogenase E1 component, theoretically possesses deacetylase activity, this possibility is low. First, in the SDS-PAGE gel of the purified recombinant protein, CobQ is the major band, with other proteins present at very low levels (less than 1/10 of CobQ). This suggests that significant deacetylation by contaminants is unlikely. Second, we purified His-tagged AhCobQ and GST-fused AhCobQ separately and tested their deacetylase activities. As shown in Figure S4 of the updated manuscript, both purified AhCobQ proteins exhibited deacetylase activity, while the negative control (purified GST protein only) did not, further supporting our conclusion that enzyme activity is not attributable to contaminating proteins (Figure S5).
(2) Without the CobB and CobQ bands, it is impossible to say the function of CobQ is a new deacetylase. To avoid this confusion, it is easy to run a new gel and stain it with anti-His antibody to show these deacetylases.
Thank you very much for your suggestion. We have performed the experiment in the comment (1) as your suggestion.
(3) The explanation about the CobB/CobQ bands are not visible is not acceptable. Because the molecular weight of the CobB and CobQ is smaller than that of BSA, it is impossible that these bands will be loss during membrane transfer.
Thank you for your valuable feedback. I completely agree that the loss of CobB and CobQ proteins during membrane transfer is unlikely due to their smaller molecular weight compared to BSA. As shown in Figure 7, the bands for CobB and CobQ are detectable in the SDS-PAGE gel but not visible on the Coomassie R-350 stained PVDF membrane.
Several factors could contribute to this issue. One possibility is that the detection sensitivity of Coomassie R-350 may be lower than that of Coomassie R-250 used in the gel. Additionally, the Western blot results using an anti-His antibody further indicate low loading amounts of CobB and CobQ proteins on the PVDF membrane (Figure 8). This suggests that the observed low levels may indeed be due to protein loss during the membrane transfer process, despite their relatively small size.
Reviewer #3 (Recommendations for the authors):
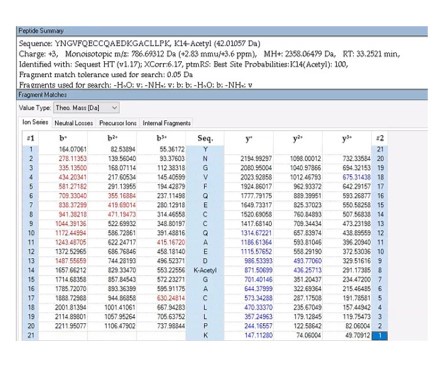
(1) I found Tables S1 and S2 in the revised manuscript. It is strange to me that the intensity of Kac-BSA+CobQ is zero, completely nothing. Typically, a portion of the acetylated peptide remains after the deacetylation reaction.
Thank you for your observation. When we report an intensity of zero, it does not imply a complete absence of signal; rather, it indicates that the signal for the target peptide is below the detectable threshold. This is likely due to the minimum cut-off setting in the MaxQuant (MQ) software, which is determined by parameters like "peptide_mass_tolerance" (as discussed in MQ user groups online, though it may not be explicitly listed in the parameters file).
In our study, we performed a deacetylase assay that demonstrated CobQ's rapid activity; for instance, it can deacetylate ICD-K388ac within just four minutes. This leads me to hypothesize that the CobQ + Kac-BSA sample may have undergone near-complete enzymatic hydrolysis during the reaction.
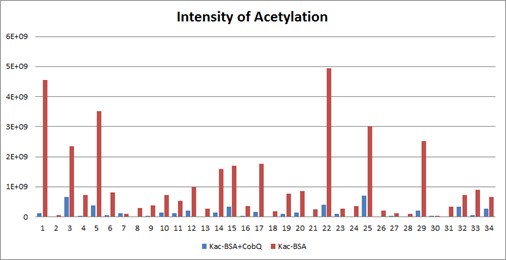
Furthermore, Table S1 in manuscript presents only a selection of the mass spectrometry results to illustrate CobQ's activity. In addition to the 15 acetylated peptides shown, there are many more (27 peptides) that exhibit significantly reduced acetylation levels without reaching zero intensity. The overall acetylation level of BSA peptides incubated with CobQ is calculated to be only 0.13 times that of Kac-BSA (Diagnostic peak: yes, peptide score: >100, Localization probability: >0.95) (Author response image 9).
Based on these findings, we believe our mass spectrometry results are reliable and effectively support our conclusions. Thank you for your understanding.
Author response image 9.
The intensities of all Kac peptides of Kac-BSA with or without AhCobQ incubation in LC MS/MS.
(2) It would be better to provide the information about ArcA and ArcA-2 as mentioned in the authors' response. It would be helpful for readers to understand that they are different proteins.
Thank you for your suggestion. In the A. hydrophila ATCC 7966 dataset, there are indeed two distinct proteins referred to as ArcA: ArcA-1, which functions as an aerobic respiration control protein, and ArcA-2, which acts as an arginine deiminase. Importantly, these two proteins do not share any sequence homology; they are only similarly named due to their acronyms. While we believe this distinction does not require extensive explanation in the current study, we appreciate your input. Additionally, in response to Reviewer 2’s feedback, we have decided to remove the Western blot result for ArcA-2 due to its poor quality in the updated manuscript.
(3) Line 409-416. Despite my comment, the citation of related papers on ICD acetylation in E. coli is still missing.
Thank you for your suggestion. It has been added and highlighted in red. (Venkat S, et al, 2018, 430(13): 1901-1911)
(4) The image resolution of Figure 3C and 3D is still bad. I could not evaluate that Kac was exactly incorporated at the target site.
Thank you for your feedback regarding the image resolution of Figures 3C and 3D. We have now displayed these figures with improved clarity, as you suggested.
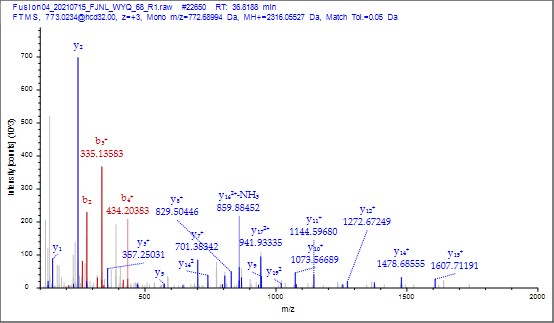
To further validate the reliability of our MS2 data, we employed Proteome Discoverer 2.4 (Thermo) to analyze the raw data and provide theoretical mass information. As shown in Author response images 10-13, the MS2 spectra and fragment match lists for both unmodified and acetylated peptides offer additional confirmation of the reliability of our mass spectrometry results.
Author response image 10.
MS2 spectrum of unmodified peptide using PD v2.4 software.
Author response image 11.
The theoretical mass of unmodified peptide by PD 2.4
Author response image 12.
MS2 spectrum of acetylated peptide using PD v2.4 software.
Author response image 13.
The theoretical mass of acetylated peptide by PD 2.4.
(5) Again, in Figure 8D, it should be shown the significance between ICD-Kac388 and ICD-Kac388+AhCobB to support the authors' conclusion that AhCobQ activates ICD by deacetylation at K388.
Thanks for your suggestion, we have updated the figure in Figure 8D in updated manuscript.
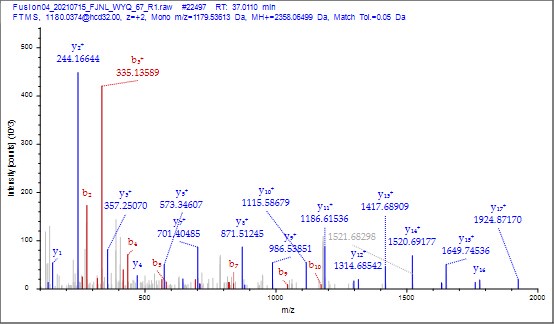
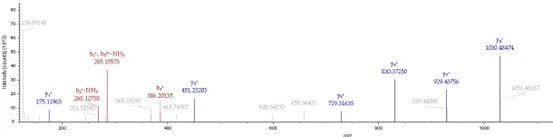
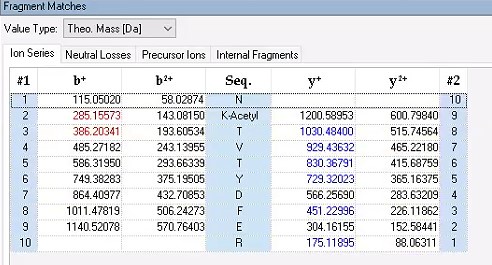
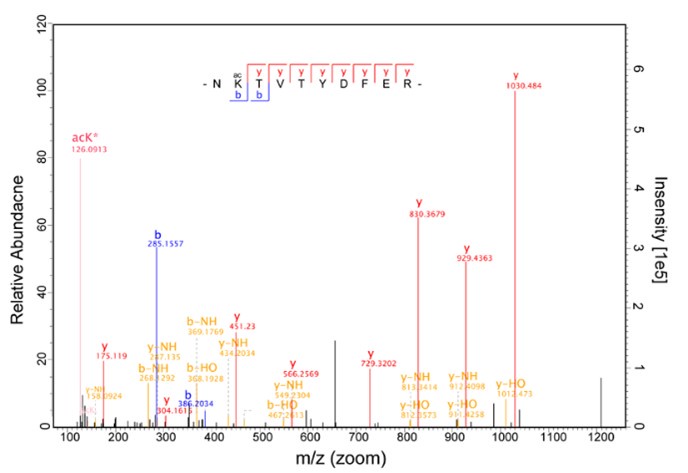
(6) It was nice that the authors presented the mass spectrum data of ICD-K388 acetylation (Figure 2 in responding letter). However, the data did not convince me that K388 is acetylated. In the figure, two b-ion peaks are detected, 285.1557 and 386.2034, which may correspond to NK (theoretical mass, 260.15) and NKT (theoretical mass, 361.20) peptides, respectively. If K388 is acetylated, an increase in the mass of 42 should be observed, but the difference between the detected and theoretical mass is 25. I also could not understand what the peak of 126.0913 mass is, indicated with acK* in red.
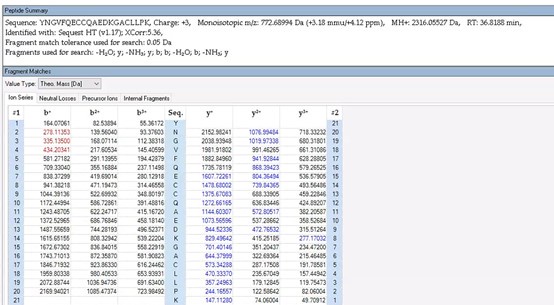
Thank you for your detailed observation. The data presented in the MS2 spectrum for ICD-K388 acetylation in Figure 2 of the previous response letter were generated using Proteome Discoverer 2.4 (PD, Thermo) to ensure accurate mass calculations. Similar to the results from MaxQuant, ICD-K388 was identified again (Author response image 14).
Regarding the b-ion peaks you mentioned, the values 285.1557 and 386.2034 correspond to NKac and NKacT peptides, respectively. The theoretical masses for these peptides are as follows: NKac (285.15 = 115.05020 + 128.095 + 42.01) and NKacT (386.20 = NKac + 101.04768). The differences between the theoretical and detected masses for the relevant b-ions (b2*-NK, b52+-NH3, and b3) are minimal, at 0.00 Da and 2.1 ppm, respectively, which is consistent with the incorporation of an NH3 group (Author response image 15).
Author response image 14.
The MS2 of ICD-K388 peptide by PD 2.4.
Author response image 15.
The theoretical mass of ICD-K388 peptide by PD 2.4.
The peak at 126.0913 m/z, indicated as acK*, represents immonium ions of ε-N-acetyllysine, which are generated during the fragmentation of acetyllysine. This diagnostic ion is widely recognized as a marker for identifying acetylated peptides (Nakayasu, et al,. A method to determine lysine acetylation stoichiometries. International journal of proteomics. 2014;2014(1):730725; Trelle et al., Utility of immonium ions for assignment of ε-N-acetyllysine-containing peptides by tandem mass spectrometry. Analytical chemistry. 2008;80(9):3422-30). Additionally, it is a default parameter in MaxQuant for identifying Kac peptides (Author response image 16).
Based on these findings, we believe the evidence supporting ICD-K388 acetylation is robust.
Author response image 16.
The default parameter in Kac peptide identification in Maxquant v1.6 software
(7) As mentioned by other reviewers, some of the figures and tables are incomplete. Some panels (ex. Figure 7C and 7D) and explanations (ex. What are lanes 1, 2, and 3 in Figure S3) are still missing.
Thank you for your suggestion. It has been added.
-

-

-

eLife assessment
In this valuable study, the authors studied a novel Zn2+- and NAD+-independent KDAC protein, AhCobQ, in Aeromonas hydrophila, which lacks homology with eukaryotic counterparts, thus underscoring its unique evolutionary trajectory within the bacterial domain. They attempt to demonstrate deacetylase activity, however, assays to detect this are still incomplete and require further refinement. The work will be of interest to microbiologists studying metabolism and post-translational modifications.
-

Reviewer #1 (Public review):
Summary:
This study by Wang et al. identifies a new type of deacetylase, CobQ, in Aeromonas hydrophila. Notably, the identification of this deacetylase reveals a lack of homology with eukaryotic counterparts, thus underscoring its unique evolutionary trajectory within the bacterial domain.
Strengths:
The manuscript convincingly illustrates CobQ's deacetylase activity through robust in vitro experiments, establishing its distinctiveness from known prokaryotic deacetylases. Additionally, the authors elucidate CobQ's potential cooperation with other deacetylases in vivo to regulate bacterial cellular processes. Furthermore, the study highlights CobQ's significance in the regulation of acetylation within prokaryotic cells.
Weaknesses:
The problem I raised has been well resolved. I have no further questions.
-

Reviewer #2 (Public review):
In recent years, lots of researchers tried to explore the existence of new acetyltransferase and deacetylase by using specific antibody enrichment technologies and high resolution mass spectrometry. Here is an example for this effort. Yuqian Wang et al. studied a novel Zn2+- and NAD+-independent KDAC protein, AhCobQ, in Aeromonas hydrophila. They studied the biological function of AhCobQ by using biochemistry method and MS identification technology to confirm it. These results extended our understanding of the regulatory mechanism of bacterial lysine acetylation modifications. However, I find this conclusion is a little speculative, and unfortunately, it also doesn't totally support the conclusion as the authors provided.
-

Reviewer #3 (Public review):
Summary:
This study reports on a novel NAD+ and Zn2+-independent protein lysine deacetylase (KDAC) in Aeromonas hydrophila, termed as AhCobQ (AHA_1389). This protein is annotated as a CobQ/CobB/MinD/ParA family protein and does not show similarity with known NAD+-dependent or Zn2+-dependent KDACs. The authors showed that AhCobQ has NAD+ and Zn2+-independent deacetylase activity with acetylated BSA by western blot and MS analyses. They also provided evidence that the 195-245 aa region of AhCobQ is responsible for the deacetylase activity, which is conserved in some marine prokaryotes and has no similarity with eukaryotic proteins. They identified target proteins of AhCobQ deacetylase by proteomic analysis and verified the deacetylase activity using site-specific Kac proteins. Finally, they showed that AhCobQ …
Reviewer #3 (Public review):
Summary:
This study reports on a novel NAD+ and Zn2+-independent protein lysine deacetylase (KDAC) in Aeromonas hydrophila, termed as AhCobQ (AHA_1389). This protein is annotated as a CobQ/CobB/MinD/ParA family protein and does not show similarity with known NAD+-dependent or Zn2+-dependent KDACs. The authors showed that AhCobQ has NAD+ and Zn2+-independent deacetylase activity with acetylated BSA by western blot and MS analyses. They also provided evidence that the 195-245 aa region of AhCobQ is responsible for the deacetylase activity, which is conserved in some marine prokaryotes and has no similarity with eukaryotic proteins. They identified target proteins of AhCobQ deacetylase by proteomic analysis and verified the deacetylase activity using site-specific Kac proteins. Finally, they showed that AhCobQ activates isocitrate dehydrogenase by deacetylation at K388.
Strengths:
The finding of a new type of KDAC has a valuable impact on the field of protein acetylation. The characters (NAD+ and Zn2+-independent deacetylase activity in an unknown domain) shown in this study are very unexpected.
Weaknesses:
(1) The characters (NAD+ and Zn2+-independent deacetylase activity in an unknown domain) shown in this study are very unexpected. To convince readers, MSMS data must be necessary to accurately detect (de)acetylation at the target site in the deacetylase activity assay. The authors showed the MSMS data in assays with acetylated BSA, but other assays only rely on western blot.
(2) They prepared site-specific Kac proteins and used them in deacetylase activity assays. Incorporation of acetyllysine at the target site should be confirmed by MSMS and shown as supplementary data.
(3) The authors imply that the 195-245 aa region of AhCobQ may represent a new domain responsible for deacetylase activity. The feature of the region would be of interest but is not sufficiently described in Figure 5. The amino acid sequence alignments with representative proteins with conserved residues would be informative. It would be also informative if the modeled structure predicted by AlphaFold is shown and the structural similarity with known deacetylases is discussed.
-

Author response:
The following is the authors’ response to the original reviews.
Public Reviews:
Reviewer #1 (Public Review):
Summary:
This study by Wang et al. identifies a new type of deacetylase, CobQ, in Aeromonas hydrophila. Notably, the identification of this deacetylase reveals a lack of homology with eukaryotic counterparts, thus underscoring its unique evolutionary trajectory within the bacterial domain.
Strengths:
The manuscript convincingly illustrates CobQ's deacetylase activity through robust in vitro experiments, establishing its distinctiveness from known prokaryotic deacetylases. Additionally, the authors elucidate CobQ's potential cooperation with other deacetylases in vivo to regulate bacterial cellular processes. Furthermore, the study highlights CobQ's significance in the regulation of acetylation within prokaryotic …
Author response:
The following is the authors’ response to the original reviews.
Public Reviews:
Reviewer #1 (Public Review):
Summary:
This study by Wang et al. identifies a new type of deacetylase, CobQ, in Aeromonas hydrophila. Notably, the identification of this deacetylase reveals a lack of homology with eukaryotic counterparts, thus underscoring its unique evolutionary trajectory within the bacterial domain.
Strengths:
The manuscript convincingly illustrates CobQ's deacetylase activity through robust in vitro experiments, establishing its distinctiveness from known prokaryotic deacetylases. Additionally, the authors elucidate CobQ's potential cooperation with other deacetylases in vivo to regulate bacterial cellular processes. Furthermore, the study highlights CobQ's significance in the regulation of acetylation within prokaryotic cells.
Weaknesses:
While the manuscript is generally well-structured, some clarification and some minor corrections are needed.
Reviewer #2 (Public Review):
In recent years, lots of researchers have tried to explore the existence of new acetyltransferase and deacetylase by using specific antibody enrichment technologies and high-resolution mass spectrometry. This study adds to this effort. The authors studied a novel Zn2+- and NAD+-independent KDAC protein, AhCobQ, in Aeromonas hydrophila. They studied the biological function of AhCobQ by using a biochemistry method and used MS identification technology to confirm it. The results extend our understanding of the regulatory mechanism of bacterial lysine acetylation modifications. However, I find their conclusion to be a little speculative, and unfortunately, it also doesn't totally support the conclusion that the authors provided. In addition, regarding the figure arrangement, lots of the supplementary figures are not mentioned, and tables are not all placed in context.
Major concerns:
- In the opinion of this reviewer, is a little arbitrary to come to the title "Aeromonas hydrophila CobQ is a new type of NAD+- and Zn2+-independent protein lysine deacetylase in prokaryotes." This should be modified to delete the "in the prokaryotes", unless the authors get new or more evidence in the other prokaryotes for the existence of the AhCobQ.
Thanks for your suggestions. " in the prokaryotes " has been deleted in the revised manuscript.
- I was confused about the arrangement of the supplementary results. There are no citations for Figures S9-S19.
Thank you very much for your suggestion. We have made revisions and highlighted in the undated manuscript.
- No data are included for Tables S1-S6.;
Dear reviewer, sorry to confuse you. We have included the Supplementary Tables in the undated manuscript.
- The load control is not all integrated. All of the load controls with whole PAGE gel or whole membrane western blot results should be provided. Without these whole results, it is not convincing to come to the conclusion that the authors have.
Dear reviewer, thanks for your suggestion. We have meticulously incorporated the complete PVDF membranes from our Western blot experiments into Supplementary Material 1. Furthermore, we have included the Coomassie Blue R-350 staining outcomes of these PVDF membranes, post-Western blot detection, as a loading control in accordance with the protocol outlined in the reference by Charlotte et al. (Journal of Proteome Research, 2011, 10:1416–1419).
- The materials & methods section should be thoroughly reviewed. It is unclear to me what exactly the authors are describing in the method. All the experimental designs and protocols should be described in detail, including growth conditions, assay conditions, purification conditions, etc.
Dear reviewer, thanks for your valuable comments. We have carefully reviewed the entire manuscript and made revisions, highlighted in red.
- Relevant information should be included about the experiments performed in the figure legends, such as experimental conditions, replicates, etc. Often it is not clear what was done based on the figure legend description.
Thank you very much for your suggestion. We have made revisions and highlighted in red.
Reviewer #3 (Public Review):
Summary:
This study reports on a novel NAD+ and Zn2+-independent protein lysine deacetylase (KDAC) in Aeromonas hydrophila, termed AhCobQ (AHA_1389). This protein is annotated as a CobQ/CobB/MinD/ParA family protein and does not show similarity with known NAD+-dependent or Zn2+-dependent KDACs. The authors show that AhCobQ has NAD+ and Zn2+-independent deacetylase activity with acetylated BSA by western blot and MS analyses. They also provide evidence that the 195-245 aa region of AhCobQ is responsible for the deacetylase activity, which is conserved in some marine prokaryotes and has no similarity with eukaryotic proteins. They identified target proteins of AhCobQ deacetylase by proteomic analysis and verified the deacetylase activity using site-specific acetyllysine-incorporated target proteins. Finally, they show that AhCobQ activates isocitrate dehydrogenase by deacetylation at K388.
Strengths:
The finding of a new type of KDAC has a valuable impact on the field of protein acetylation. The characters (NAD+ and Zn2+-independent deacetylase activity in an unknown domain) shown in this study are very unexpected.
Weaknesses:
(1) As the characters of AhCobQ are very unexpected, to convince readers, MSMS data would be needed to exactly detect deacetylation at the target site in deacetylase activity assays. The authors show the MSMS data in assays with acetylated BSA, but other assays only rely on western blot.
(2) They prepared site-specific Kac proteins and used them in deacetylase activity assays. The incorporation of acetyllysine at the target site needs to be confirmed by MSMS and shown as supplementary data.
(3) The authors imply that the 195-245 aa region of AhCobQ may represent a new domain responsible for deacetylase activity. The feature of the region would be of interest but is not sufficiently described in Figure 5. The amino acid sequence alignments with representative proteins with conserved residues would be informative. It would be also informative if the modeled structure predicted by AlphaFold is shown and the structural similarity with known deacetylases is discussed.
Recommendations for the authors:
Reviewer #1 (Recommendations For The Authors):
(1) The protein molecules of AhCobB and AhCobQ are greater than 45 kDa. But the gene sequences don't seem to match. Please explain.
We are sorry to confuse you. The vector used for the purification of CobB and CobQ in the manuscript is pET-32a, which carries the TrxA fusion protein and is approximately 20kDa in size. Therefore, the final molecular weight of recombinant AhCobB and AhCobQ is 48.3(28.3+ ~20kDa) and 49.8 (29.8+ ~20kDa), respectively.
(2) Figure 7: The gels look very smeary. Please explain.
Dear esteemed reviewer, in our study, we have meticulously crafted recombinant site-specific Kac proteins utilizing an innovative two-plasmid system, grounded on the seminal work published in Nature Chemical Biology (2017, 13(12): 1253-1260), which introduced the genetic encoding of Nᵋ-acetyllysine into recombinant proteins. However, we have encountered a prevalent challenge—the occurrence of protein truncation due to premature translation termination at the reassigned codon. This phenomenon not only diminishes protein yields, as highlighted in ChemBioChem (2017, 18(20): 1973-1983), but also plagues many recombinant proteins with a troublesome backdrop in Western Blot (WB) outcomes.
Despite our rigorous approach, involving at least two independent repetitions for WB analysis of site-specific Kac proteins, yielding consistent results, we acknowledge that the overall quality of these WB assays remains suboptimal. This variability is inherently tied to the intrinsic properties of the target proteins themselves. Illustratively, the WB outcomes for proteins such as ENO and ICD exhibit notable differences in quality across biological replicates, emphasizing the complexity and nuances involved in this process.
Thus, while our methodology remains robust and reproducible, we are mindful of the limitations imposed by the nature of the proteins under investigation and strive to continually refine our approaches to mitigate these challenges.
(3) To ensure that the phenotype shown in Figure 1 is not due to polar effects, results of supplementing complementary strains should be provided.
Thank you for your suggestion. We have constructed a complement strain and tested the bacterial migration ability. As shown in the Figure S1, the complement strain does not affect the physiological phenotype mentioned above.
(4) The caption to Figure 8 includes * and *** to indicate significance levels, but only *** appears in the picture.
Thank you for your suggestion. It has been modified and highlighted in red.
(5) Has the mechanistic role of lysine 388 in ICD been characterized?
Thank you for your invaluable professional insights. Indeed, the acetylation sites of ICD have been established to exert a significant influence on its enzymatic activity. Sumana Venkat et al., in their seminal work published in the Journal of Molecular Biology (2018, 430(13): 1901-1911), convincingly demonstrated that the acetylation of specific lysine residues—K100, K230, K55, and K350—in ICD proteins from E. coli serves as a negative regulatory mechanism for enzyme activity. Intriguingly, the functional implications of the Kac modification on K387 (corresponding to the K388 site in ICD from A. hydrophila ATCC 7966, as featured in this manuscript) remain an uncharted territory.
Our experimental endeavors have illuminated that the K388 site of ICD in A. hydrophila holds the potential to modulate enzymatic activity and is under the regulatory influence of AhCobQ.
(6) The format of the references is not uniform enough, for example, some journal names are abbreviated, and some are not, please check and correct.
Thank you for your suggestion. It has been modified and highlighted in red.
(7) Page 23, line 13, gene not expressed in italics, please correct.
Thank you for your suggestion. It has been modified and highlighted in red.
(8) Figure S8 does not appear to match the gene size.
We are sorry to confuse you. The vector used for the purification of recombinant protein in the manuscript is pET-32a, which carries the TrxA fusion protein and is approximately 20kDa in size. Therefore, the final molecular weight of recombinant protein is 25.5(5.5+ ~20kDa).
(9) The format of the two figures in Figure S10 is not uniform.
Thank you for your suggestion. It has been modified and highlighted in red.
Reviewer #2 (Recommendations For The Authors):
Minor concerns:
L147, L177 - Please arrange the results as they are shown in the content sequentially. For example, rename Figure S2 with Figure S1.
Thank you for your suggestion. It has been modified and highlighted in red.
L174 Figure 2D - There is no big change in the acetylation between the wild type and ahcobQ mutant from Figure 2D, but the ahcobB mutant is.
I am extremely grateful for your insightful comment. As clearly depicted in the right panel of Figure 2D, the overall Kac protein levels in both the ahcobQ and ahcobB knockout strains exhibit a marked elevation compared to the wild-type strain, despite equivalent loading of total cellular proteins (the left panel of Figure 2D). Notably, this increase is particularly pronounced among proteins with a molecular weight below 35 kDa. We wholeheartedly concur with your perspective that the deletion of ahcobB leads to a more substantial enhancement in Kac protein levels, suggesting CobB may play a pivotal role in regulating a broader spectrum of acetylated proteins or Kac sites. This hypothesis is further strengthened by subsequent mass spectrometry analyses, which lend additional credence to our shared understanding.
L174-187, L795 - Please show the whole membrane (or PAGE gel) of the loading control of CobB, and CobQ, except for the Kac-BSA.
Dear esteemed reviewer, we have thoroughly revised our submission to include the full western blot (WB) membrane for all figures and supplementary figures within the updated Supplementary Material 1. Additionally, we would like to clarify a few crucial points to ensure transparency and accuracy.
Firstly, in Figure 2D, we present WB results solely pertaining to whole-cell samples from cobB or cobQ mutant strains. Consequently, these findings do not directly correlate with recombinant CobB or CobQ proteins.
Secondly, the objective of Figure 2 is to validate the lysine deacetylase activity of AhCobQ protein through a qualitative, rather than quantitative, experimental approach. Hence, the crucial loading control lies in the amount of Kac-BSA, rather than CobB or CobQ. Prior to conducting the in vitro deacetylase assay, we ensured equal protein concentrations of purified CobB or CobQ using BCA assay, adhering to the protocol's specified deacetylase-to-Kac-BSA loading ratio of 1:5. However, this ratio renders the deacetylase (CobB or CobQ) undetectable on Coomassie Blue R-350-stained blots or WB membranes (as detailed in the whole WB membrane in Supplementary Material 1).
To reinforce our observations, we reiterated the analysis of protein samples by subjecting them once again to SDS-PAGE, maintaining the same loading quantity as utilized in the preceding western blotting experiment shown in Figure 2E. As Author response image 1 clearly illustrates, the CobB/CobQ bands are indeed discernible, albeit they exhibit significantly fainter intensities when compared to the Kac-BSA bands. Notably, upon reviewing the full strained PVDF membrane presented in Supplementary Material 1, we find that the CobB/CobQ bands are not readily visible. This observation can be attributed to the potential loss of proteins during the transfer process from SDS-PAGE to the PVDF membrane.
Author response image 1.
The SDS-PAGE gel displayed the loading amounts of Kac-BSA and CobB/CobQ.
Furthermore, recognizing the potential for confusion given the similar molecular weights of CobB (257aa) and CobQ (264aa, excluding fusion tags), we conducted a comparative analysis of deacetylase activity between His-tagged and GST-fused recombinant CobQ proteins. Encouragingly, both variants exhibited deacetylase activity (as presented in Figure S5 of the revised manuscript), thereby excluding any influence from nonspecific proteins that might have contaminated the purification process.
We hope these clarifications and additions to our submission address your concerns and enhance the overall quality of our work. Thank you for your valuable time and consideration.
- Could you provide the raw data of these anti-acetylation western blot results?
Thank you very much for your suggestion. The raw results have been uploaded in the supplementary materials.
- According to the loading control, the protein quantity of BSA is very big, however, why is the acetylation of Kac-BSA relatively low? Is it consistent between the western blot and loading control?
Thank you very much for your suggestion, first of all, all the western blot and loading control in the manuscript are the same membrane, and the specific method is described in "Western blot". Therefore, there is no possibility that the western blot and loading control do not correspond. Secondly, not every site of BSA has acetylation modifications, and the amount of modifications at each site is also different, so there will be a large amount of protein but a small amount of acetylation.
Figure 2C - Could the Dot blot experiment be described in detail in the Methods part?
Thank you for your suggestion. It has been added and highlighted in red.
Figure 2C&2D - Please provide the anti-acetylation antibody information.
Thank you for your suggestion. It has been added and highlighted in red.
Figure 2E - It is confusing why the acetylation of Kac-BSA is higher than adding NAD+ with CobB? But only CobB can deacetylate the Kac-BSA without NAD+?
We are sorry to confuse you. The information in the figure is incorrect. For somehow, we provided the uncorrected version, and we have revised it in the undated manuscript.
Figure 2F - The control of this experiment should include the NAM, CobB, and NAM+CobB. Similar to 2E, it also should include NAD, CobB, and NAD+CobB, respectively. Same with 2H.
We are sorry to confuse you. The intent of Figure 2F is to further confirm that AhCobQ is different from AhCobQ and can remove the acetylation modification of BSA without relying on NAD+, so NAD+ was added to this group of experiments. We have revised the manuscript to add details about the experiments.
L178 Figure S1C - One question about the protein AhAcuC. From the PCR results, it is larger than ahcobB and ahcobQ, however, why is the protein AhAcuC smaller than them?
We are sorry to confuse you. The images in the original manuscript may have had some errors in protein size due to different PAGE gels. We have re-run the gels and replaced them in the manuscript in the Supplementary Figure S3 in revised manuscript.
- All the proteins are expressed and purified from E.coli BL21(DE3). How did you avoid the pollution of the deacetylase from the E.coli? There is no control over it in your experiment. Without this control, it is not easy to come to the conclusion that the deacetylation is from the AhCobQ but not from the pollution from the protein purification.
In response to your inquiry, we have conducted a meticulous comparative analysis of the deacetylase activity exhibited by both His-tagged and GST-fused recombinant AhCobQ proteins. Reassuringly, our findings reveal that both variants possess robust deacetylase activity, as clearly demonstrated in Figure S5 of the revised manuscript. Furthermore, to ensure the rigor of our experiments, we employed GST protein purified from E.coli strains as a negative control in Figure S8. The Western blot (WB) results conclusively demonstrate that GST protein alone lacks deacetylase activity, thereby reinforcing the authenticity of our findings and effectively mitigating any concerns regarding potential interference from nonspecific proteins during the purification process.
L190 - Could you provide the raw data for Table S1?
Thank you very much for your suggestion. The raw MS data were deposited in the public ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD038735 or IPX0005366000(iProx database). We also uploaded the analysis results in Table S1 and Supplementary material 2.
- I am not an expert on MS. I have one question about the MS results. Why there is no peak for the CobB or CobQ as they add to the reaction system?
Thank you for your insightful question. To clarify, the Kac peptides identified from Kac-BSA, as presented in Table S1, were meticulously selected for the purpose of enhancing their display and facilitating interpretation. The comprehensive raw mass spectrometry (MS) data, along with detailed analytical outcomes, have been diligently deposited within the ProteomeXchange Consortium, specifically through the PRIDE partner repository, under the dataset identifier PXD038735 or alternatively accessible via the iProx database under IPX0005366000. The analysis results also included in the Table S1 and Supplementary material 2.
Furthermore, it is crucial to note that in this study, we utilized Bovine serum albumin (BSA) as the foundational database for our MS searches. Consequently, the absence of CobB or CobQ proteins in our MS results stems from the inherent focus on BSA and the specific experimental design, which did not encompass the detection of these particular proteins.
We appreciate your attention to these details and hope this clarification addresses your query.
L189-L206 - Based on the results here, the function of CobB and CobQ overlaps on the same STDKac peptides.
Dear esteemed reviewer, our mass spectrometry (MS) analysis has revealed an intriguing finding: CobB and CobQ indeed function on the same STDKac peptide, suggesting a potential collaboration among distinct deacetylases in regulating protein function. This observation is further corroborated by our subsequent quantitative Kac proteomics results, which were obtained from three deacetylase mutants. These results underscore the possibility that CobB, CobQ, and AcuC possess both unique and overlapping protein substrates, reinforcing our hypothesis that multiple deacetylases work in concert to modulate protein activity.
- Do you assay the Km and Kcat about the CobQ by using Kac-BAS as the substrate by comparing with AhCobB?
Dear reviewer, thanks for your professional suggestion. In accordance with your guidance, we diligently attempted to analyze the Km or Kcat values of CobQ during its incubation with the substrate Kac-BSA using LC-MS/MS, repeating the process twice. However, to our disappointment, our current experimental platform has been unable to detect any discernible metabolites. We suspect that this may stem from operational proficiency challenges, as even our positive control experiment involving CobB incubation has failed to yield satisfactory results.
Given our uncertainty regarding the root cause of these issues, coupled with the suggestion from experts that the LC column might be a contributing factor except for skill, we have decided against repeating the experiments at this juncture. Nonetheless, we would like to assure you that we have rigorously validated the deacetylase activity of CobQ proteins through mass spectrometry, as detailed in our manuscript.
Furthermore, I am delighted to share that our preliminary findings have sparked interest among other research teams. In fact, one such group, upon reading our preprint, has independently tested the activity of CobQ and uncovered an additional intriguing function. We are actively exploring the possibility of collaborating with this team to delve deeper into the research and, hopefully, in the future, conduct a more refined analysis of the Km and Kcat of CobQ.
L214- Same question with Figures 2E-2H. Could you provide the whole page gel about the loading control? I want to know the quantity of the AhCobQ in this experiment except for the Kac-BSA. To tell the truth, the quantity of BSA is too much in the deacetylation reaction system to be able to tell its deacetylation activity in vitro.
Thank you very much for your suggestion. The raw data has been uploaded in the supplementary materials and the clarification is similar with above mentioned.
L217 - There might be a wrong citation of Figure S2 here.
Thank you for your suggestion. It has been corrected.
L244-250, Figure 6A - Are there 47, not 46 Kac proteins?
Thank you for your suggestion. It has been corrected.
- Are there nineteen, not nine increased Kac peptides common between the ΔahcobQ and ΔahacuC strains?
Thank you for your suggestion. It has been corrected.
- Are there ten, not six increased Kac peptides common between the ΔahcobQ and ΔahcobB strains?
Thank you for your suggestion. It has been corrected.
- Are there 69, not 65 increased Kac peptides common between the ΔahcobB and ΔahacuC strains?
Thank you for your suggestion. It has been corrected.
- Where is the raw data for Table S2?
Thank you very much for your suggestion. The raw data has been uploaded in the supplementary materials.
Figure 6B - Are there 52, not 51 Kac peptides?
Thank you for your suggestion. It has been corrected.
L272 - Why do you choose these 11 target proteins? There is no description of this background in the context.
We have opted to prioritize these proteins for subsequent validation, as their Kac levels exhibit a notable upregulation in the ΔahcobQ strain, potentially indicating their role as protein substrates for AhCobQ. We will incorporate this clarification into the revised manuscript to ensure clarity and comprehensiveness.
L277 - Figure S6 - Please show the whole PAGE gel about the loading control.
Dear esteemed reviewer, we sincerely apologize for any confusion our previous presentation may have caused. We would like to clarify that the bottom panel of Figure S6 depicts a Coomassie Blue R-350 stained whole PVDF membrane, rather than a PAGE gel, as may have been mistakenly inferred. To facilitate a comprehensive understanding, we have included the entire stained PVDF membranes in Supplementary Material 1.
As we have previously elaborated, the recombinant His-tagged or GST-fused AhCobQ proteins were not as discernible on the PVDF membrane due to a relatively lower loading amount compared to that of Kac-BSA.
-There might be a wrong citation in Figure S6. As you mentioned in the context, you expressed and purified 11 proteins and then tested their acetylation background.
Thank you for your suggestion. It has been corrected.
L280 - Figure S7 -The label of the Figure should be modified for the ATP.
Thank you for your suggestion. It has been modified.
- How did you do the experiment for 0h of ATP? There is no description of it in the Methods.
Thank you for your suggestion. It has been added.
- Please show the whole PAGE gel about the loading control.
Thank you very much for your suggestion. The whole PAGE gel has been uploaded in the supplementary materials.
L282 - Figure 7 - Please show the whole PAGE gel about the loading control.
Dear esteemed reviewer, we sincerely apologize for any confusion our previous presentation may have caused. We would like to clarify that the bottom panel of Figure S6 depicts a Coomassie Blue R-350 stained whole PVDF membrane, rather than a PAGE gel, as may have been mistakenly inferred. To facilitate a comprehensive understanding, we have included the entire stained PVDF membranes in Supplementary Material 1.
- Please adjust the font size of "A" and "B".
Thank you for your suggestion. It has been adjusted.
Figure 7A - The anti-acetylation Western blot here does not look good. All the western blots here should be re-done.
Dear reviewer, the recombinant site-specific Kac proteins were constructed by two-plasmid system based on genetically encoding Nᵋ-acetyllysine in recombinant proteins in this study (Nature chemical biology, 2017, 13(12): 1253-1260). However, a common problem experienced is protein truncation arising from translation termination at the reassigned codon, lowering protein yields (ChemBioChem, 2017, 18(20): 1973-1983), and leading to a dirty background of WB results in many recombinant proteins. Although we did perform at least two times independent repeats for site-specific Kac protein WB and got similar results, the WB quality of site-specific Kac proteins are general poor and that depend on the properties of target proteins. For example, the WB results of ENO and ICD can display considerable qualities in different biological repeats.
- Why did you choose the PAGE gel but not the anti-His Western blot as the loading control?
Thank you very much for your suggestion. Labeling antibodies is a very effective loading control. However, in order to ensure the accuracy of the data, both the experimental data and loading control in this manuscript are required to be reflected on the same membrane. If His tags are used, the membrane will be washed repeatedly for secondary color development. Based on the fact that acetylation modification is already difficult for color development, this will greatly affect the quality of the results presented. Meanwhile, while ensuring consistent protein levels, we believe that changes in acetylation modifications can also explain the issue. Therefore, you choose the PAGE gel but not the anti-His Western blot as the loading control.
L278 - Where are the results of the site-specific lysine acetylation of the target protein by using two-plasmid-based system of genetically encoded Nε-acetyllysine. Usually, there will be a shift when it is full acetylated by compared with the wild-type protein.
Sorry for the confusion caused. As the size of the acetyl group is only about 40.6Da, which is thousands of times smaller than the size of the protein, the changes in size of the protein before and after modification cannot be seen with the naked eye.
L287 - Where is Figure 7C?
We are sorry to confuse you. It has been corrected.
- Here the citation might be Figure 7A but not Figure 7B.
Thank you for your suggestion. It has been corrected.
L290 - It is difficult to read here, please rearrange this Figure S8. There is no useful label.
Thank you for your suggestion. It has been corrected.
- The citation of Figure S8 is wrong.
Thank you for your suggestion. It has been corrected.
- For Figure S8, please add the label on the figure. And add anti-GST western blot as well. Because the GST is about 26KD, why are the purified recombinant truncated proteins (GST-fusion) so small?
Sorry for the inconvenience caused. The truncated fragment used for recombinant purification in Figure S8 is very small, and when converted to protein, it is approximately between 1-5kDa. Therefore, the resulting protein is also very small.
- Why there are two Figure S8 in the supplemental materials?
We are sorry to confuse you. It has been corrected.
L293 - Where is Figure 7D?
We are sorry to confuse you. It has been corrected.
L297-313 - Please provide the MS result of the ICDK388?
Author response image 2.
The mass spectrum of Kac modification on ICD protein at K388 site.
Dear reviewer, we are pleased to present the mass spectrum data pertaining to the Kac modification at the K388 site of the ICD protein in ΔahcobQ strain in Figure2 in this responding letter. It is important to clarify that, while we have not directly validated the Kac status of site-specific lysine acetylation at the recombinant ICD K388 site through mass spectrometry (MS) in this particular study, we have strong reasons to believe in its specificity.
Firstly, our confidence stems from the well-established and rigorously validated two-plasmid system methodology for site-directed acetylation modification. This approach has been successfully employed in modifying diverse and specific sites across various proteins, as evidenced by the pioneering work of David et al. in Nature Chemical Biology (2017, 13(12), 1253-1260).
Secondly, we have taken meticulous measures to ensure the accuracy and reliability of our findings. This includes double-checking our PCR primers and DNA sequencing for the genetic code expansion technology employed. Furthermore, we have included control experiments utilizing proteins that were not subjected to site-directed acetylation (ICD), as detailed in Figure 8A in revised manuscript, thereby providing an additional layer of validation and reinforcing the robustness of our results.
We believe that these two lines of evidence, combined with our rigorous experimental design and execution, provide a solid foundation for our conclusion regarding the specific acetylation of the K388 site in ICD.
- Please provide the whole PAGE gel of loading control. Or other anti-His results?
Dear esteemed reviewer, we sincerely apologize for any confusion our previous presentation may have caused. We would like to clarify that the bottom panel of Figure S6 depicts a Coomassie Blue R-350 stained whole PVDF membrane, rather than a PAGE gel, as may have been mistakenly inferred. To facilitate a comprehensive understanding, we have included the entire stained PVDF membranes in Supplementary Material 1.
- Do you have site-specific antibody of ICDK388? It should be better to identify the ICDK388 with site-specific anti-acetylation antibody.
Thank you for your insightful suggestion. We fully concur that a site-specific antibody targeting ICDK388 would be an optimal tool to elucidate the impact of CobQ on the acetylation status (Kac) of this protein. Unfortunately, we are currently without such an antibody due to the intricate and time-consuming process of its production, which also requires rigorous validation to ensure specificity. Furthermore, the cost associated with its development is considerable.
To address this limitation, in the present manuscript, we have innovatively employed a two-plasmid system for site-directed acetylation modification of ICDK388. This method, which has been extensively validated and utilized in modifying diverse specific sites (David et al., Nature Chemical Biology, 2017, 13(12), 1253-1260), allowed us to precisely manipulate the acetylation status of our target protein. Additionally, we incorporated control experiments using proteins that were not subjected to site-directed acetylation, as depicted in Figure 8A in revised manuscript, thereby reinforcing the robustness and reliability of our findings.
- Please give some background information about K388 site of ICD in the context.
Thank you for your suggestion. It has been added.
L484 - Could you provide the reference for this assay method "Protein deacetylation assay in vitro"?
Thank you for your suggestion. The work published in science 327, 1004 (2010) and Nat. Protoc.5, 1583-1595.
L490 - There is no detailed information about the growh condition for the quantitative acetylome analysis. Without these information, the proportion of the Kac peptides doesn't make any sense.
Thank you for your suggestion. It has been added.
L531 - Insert one line before the paragraph of Western blot.
Thank you for your suggestion. It has been inserted.
Reviewer #3 (Recommendations For The Authors):
Tables S1 and S2 are missing. I could not fully understand the manuscript without them.
We are sorry to confuse you.The data has been uploaded in the supplementary materials.
Line 130. The gene IDs of AhCobB and AhAcuC should be presented.
Thank you for your suggestion. It has been presented.
Line 285. What is different between ArcA and ArcA-2? Please clarify.
Thank you for your suggestion. ArcA is aerobic respiration control protein ArcA, gene name AHA_3026 (https://www.uniprot.org/uniprotkb/A0KMM9/entry). ArcA-2 is arginine deiminase, which gene name is AHA_4093 (https://www.uniprot.org/uniprotkb/A0KQG6/entry). Therefore, they are different proteins according to Uniport annotation.
Line 303. 8further, a bug?
We are sorry to confuse you. It has been corrected.
Line 412-416. The related papers on ICD acetylation in E. coli should be cited.
We are sorry to confuse you. It has been added.
Line 478. Not in vivo but in vitro?
Sorry to confuse you. It should be in vitro. We have revised in the updated manuscript.
Figure 3C and 3D. The image resolution is bad. The figures should be improved so that readers to know easily that Kac is exactly incorporated at the target site.
Thank you for your suggestion. It has been corrected.
Figure 4B. The amino acid residues of the whole AhCobB should be 1-264 aa.
Thank you for your suggestion. It has been corrected.
Figure 8. It would be better to use the same colors between panels C and D. It should be shown the significance between ICD-Kac388 and ICD-Kac388+AhCobB to support the authors' conclusion that AhCobQ activates ICD by deacetylation at K388.
Thank you for your suggestion. It has been adjusted.
-

-

eLife assessment
This valuable study describes a new type of NAD+ and Zn2+-independent protein lysine deacetylase in prokaryotes. These results extend the understanding of regulatory mechanisms related to bacterial lysine acetylation modifications however, the experimental evidence is incomplete and does not fully support the conclusions made. The work will be of interest to microbiologists studying metabolism and post-translational modifications.
-

Reviewer #1 (Public Review):
Summary:
This study by Wang et al. identifies a new type of deacetylase, CobQ, in Aeromonas hydrophila. Notably, the identification of this deacetylase reveals a lack of homology with eukaryotic counterparts, thus underscoring its unique evolutionary trajectory within the bacterial domain.
Strengths:
The manuscript convincingly illustrates CobQ's deacetylase activity through robust in vitro experiments, establishing its distinctiveness from known prokaryotic deacetylases. Additionally, the authors elucidate CobQ's potential cooperation with other deacetylases in vivo to regulate bacterial cellular processes. Furthermore, the study highlights CobQ's significance in the regulation of acetylation within prokaryotic cells.
Weaknesses:
While the manuscript is generally well-structured, some clarification and some …
Reviewer #1 (Public Review):
Summary:
This study by Wang et al. identifies a new type of deacetylase, CobQ, in Aeromonas hydrophila. Notably, the identification of this deacetylase reveals a lack of homology with eukaryotic counterparts, thus underscoring its unique evolutionary trajectory within the bacterial domain.
Strengths:
The manuscript convincingly illustrates CobQ's deacetylase activity through robust in vitro experiments, establishing its distinctiveness from known prokaryotic deacetylases. Additionally, the authors elucidate CobQ's potential cooperation with other deacetylases in vivo to regulate bacterial cellular processes. Furthermore, the study highlights CobQ's significance in the regulation of acetylation within prokaryotic cells.
Weaknesses:
While the manuscript is generally well-structured, some clarification and some minor corrections are needed.
-

Reviewer #2 (Public Review):
In recent years, lots of researchers have tried to explore the existence of new acetyltransferase and deacetylase by using specific antibody enrichment technologies and high-resolution mass spectrometry. This study adds to this effort. The authors studied a novel Zn2+- and NAD+-independent KDAC protein, AhCobQ, in Aeromonas hydrophila. They studied the biological function of AhCobQ by using a biochemistry method and used MS identification technology to confirm it. The results extend our understanding of the regulatory mechanism of bacterial lysine acetylation modifications. However, I find their conclusion to be a little speculative, and unfortunately, it also doesn't totally support the conclusion that the authors provided. In addition, regarding the figure arrangement, lots of the supplementary figures are …
Reviewer #2 (Public Review):
In recent years, lots of researchers have tried to explore the existence of new acetyltransferase and deacetylase by using specific antibody enrichment technologies and high-resolution mass spectrometry. This study adds to this effort. The authors studied a novel Zn2+- and NAD+-independent KDAC protein, AhCobQ, in Aeromonas hydrophila. They studied the biological function of AhCobQ by using a biochemistry method and used MS identification technology to confirm it. The results extend our understanding of the regulatory mechanism of bacterial lysine acetylation modifications. However, I find their conclusion to be a little speculative, and unfortunately, it also doesn't totally support the conclusion that the authors provided. In addition, regarding the figure arrangement, lots of the supplementary figures are not mentioned, and tables are not all placed in context.
Major concerns:
-In the opinion of this reviewer, is a little arbitrary to come to the title "Aeromonas hydrophila CobQ is a new type of NAD+- and Zn2+-independent protein lysine deacetylase in prokaryotes." This should be modified to delete the "in the prokaryotes", unless the authors get new or more evidence in the other prokaryotes for the existence of the AhCobQ.
-I was confused about the arrangement of the supplementary results. There are no citations for Figures S9-S19.
-No data are included for Tables S1-S6.
-The load control is not all integrated. All of the load controls with whole PAGE gel or whole membrane western blot results should be provided. Without these whole results, it is not convincing to come to the conclusion that the authors have.
-The materials & methods section should be thoroughly reviewed. It is unclear to me what exactly the authors are describing in the method. All the experimental designs and protocols should be described in detail, including growth conditions, assay conditions, purification conditions, etc.
-Relevant information should be included about the experiments performed in the figure legends, such as experimental conditions, replicates, etc. Often it is not clear what was done based on the figure legend description.
-

Reviewer #3 (Public Review):
Summary:
This study reports on a novel NAD+ and Zn2+-independent protein lysine deacetylase (KDAC) in Aeromonas hydrophila, termed AhCobQ (AHA_1389). This protein is annotated as a CobQ/CobB/MinD/ParA family protein and does not show similarity with known NAD+-dependent or Zn2+-dependent KDACs. The authors show that AhCobQ has NAD+ and Zn2+-independent deacetylase activity with acetylated BSA by western blot and MS analyses. They also provide evidence that the 195-245 aa region of AhCobQ is responsible for the deacetylase activity, which is conserved in some marine prokaryotes and has no similarity with eukaryotic proteins. They identified target proteins of AhCobQ deacetylase by proteomic analysis and verified the deacetylase activity using site-specific acetyllysine-incorporated target proteins. Finally, …
Reviewer #3 (Public Review):
Summary:
This study reports on a novel NAD+ and Zn2+-independent protein lysine deacetylase (KDAC) in Aeromonas hydrophila, termed AhCobQ (AHA_1389). This protein is annotated as a CobQ/CobB/MinD/ParA family protein and does not show similarity with known NAD+-dependent or Zn2+-dependent KDACs. The authors show that AhCobQ has NAD+ and Zn2+-independent deacetylase activity with acetylated BSA by western blot and MS analyses. They also provide evidence that the 195-245 aa region of AhCobQ is responsible for the deacetylase activity, which is conserved in some marine prokaryotes and has no similarity with eukaryotic proteins. They identified target proteins of AhCobQ deacetylase by proteomic analysis and verified the deacetylase activity using site-specific acetyllysine-incorporated target proteins. Finally, they show that AhCobQ activates isocitrate dehydrogenase by deacetylation at K388.
Strengths:
The finding of a new type of KDAC has a valuable impact on the field of protein acetylation. The characters (NAD+ and Zn2+-independent deacetylase activity in an unknown domain) shown in this study are very unexpected.
Weaknesses:
(1) As the characters of AhCobQ are very unexpected, to convince readers, MSMS data would be needed to exactly detect deacetylation at the target site in deacetylase activity assays. The authors show the MSMS data in assays with acetylated BSA, but other assays only rely on western blot.
(2) They prepared site-specific Kac proteins and used them in deacetylase activity assays. The incorporation of acetyllysine at the target site needs to be confirmed by MSMS and shown as supplementary data.
(3) The authors imply that the 195-245 aa region of AhCobQ may represent a new domain responsible for deacetylase activity. The feature of the region would be of interest but is not sufficiently described in Figure 5. The amino acid sequence alignments with representative proteins with conserved residues would be informative. It would be also informative if the modeled structure predicted by AlphaFold is shown and the structural similarity with known deacetylases is discussed.
-