Elimination of subtelomeric repeat sequences exerts little effect on telomere essential functions in Saccharomyces cerevisiae
Curation statements for this article:-
Curated by eLife
eLife assessment
This important study advances our understanding of the biological significance of the DNA sequence adjacent to telomeres. The data presented convincingly demonstrate that subtelomeric repeats are non-essential and have a minimal, if any, role in maintaining telomere integrity of budding yeast. The work will be of interest to the telomere community specifically and the genome integrity community more broadly.
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Telomeres, which are chromosomal end structures, play a crucial role in maintaining genome stability and integrity in eukaryotes. In the baker’s yeast Saccharomyces cerevisiae , the X- and Y’-elements are subtelomeric repetitive sequences found in all 32 and 17 telomeres, respectively. While the Y’-elements serve as a backup for telomere functions in cells lacking telomerase, the function of the X-elements remains unclear. This study utilized the S. cerevisiae strain SY12, which has three chromosomes and six telomeres, to investigate the role of X-elements (as well as Y’-elements) in telomere maintenance. Deletion of Y’-elements (SY12 YΔ ), X-elements (SY12 XYΔ+Y ), or both X- and Y’-elements (SY12 XYΔ ) did not impact the length of the terminal TG 1-3 tracks or telomere silencing. However, inactivation of telomerase in SY12 YΔ , SY12 XYΔ+Y , and SY12 XYΔ cells resulted in cellular senescence and the generation of survivors. These survivors either maintained their telomeres through homologous recombination-dependent TG 1-3 track elongation or underwent microhomology-mediated intra-chromosomal end-to-end joining. Our findings indicate the non-essential role of subtelomeric X- and Y’-elements in telomere regulation in both telomerase-proficient and telomerase-null cells and suggest that these elements may represent remnants of S. cerevisiae genome evolution. Furthermore, strains with fewer or no subtelomeric elements exhibit more concise telomere structures and offer potential models for future studies in telomere biology.
Article activity feed
-

-

-

-

Author Response
The following is the authors’ response to the previous reviews.
Reviewer #1
The authors provided experimental data in response to my comments/suggestions in the revision. Overall, most points were appropriate and satisfactory, but some issues remain.
(1) It is not fully addressed how atypical survivors are generated independently of Rad52-mediated homologous recombination.
The newly provided data indicate that the formation of atypical telomeres is independent of the Rad52 homologous recombination pathway.
"The atypical telomeres clones exhibit non-uniform telomere pattern", but the TG-hybridized signals after XhoI digestion are clear and uniform.
"Atypical telomere" clones may carry circular chromosomes embedded with short TG repeats, rather than linear chromosomes. In other words, atypical telomeres may …
Author Response
The following is the authors’ response to the previous reviews.
Reviewer #1
The authors provided experimental data in response to my comments/suggestions in the revision. Overall, most points were appropriate and satisfactory, but some issues remain.
(1) It is not fully addressed how atypical survivors are generated independently of Rad52-mediated homologous recombination.
The newly provided data indicate that the formation of atypical telomeres is independent of the Rad52 homologous recombination pathway.
"The atypical telomeres clones exhibit non-uniform telomere pattern", but the TG-hybridized signals after XhoI digestion are clear and uniform.
"Atypical telomere" clones may carry circular chromosomes embedded with short TG repeats, rather than linear chromosomes. In other words, atypical telomeres may differ from telomeres, the ends of chromosomes. Is atypical telomere formation dependent on NHEJ? Given that "two chromosomes underwent intra-chromosomal fusions" (Line 248), are atypical telomere clones detected frequently in SY13 cells containing two chromosomes?
We thank the reviewer’s questions. Frankly, we have not been able to determine the chromosome structures in these so-called "atypical survivors". As we mentioned in the manuscript, there could be mixed telomere structures, e.g. TG tract amplification, intro-chromosome telomere fusion and inter-chromosome telomere fusion. Worse still, these 'atypical survivors' may not have maintained a stable genome, and their karyotype may have undergone stochastic changes during passages. To avoid misunderstanding, we change the term "atypical" to "uncharacterized" in the revised manuscript.
We have previously shown that deletion of YKU70 does not affect MMEJ-mediated intra-chromosome fusion in single-chromosome SY14 cdc13Δ cells (Wu et al., 2020). In SY12 cells, double knockout of TLC1 and YKU resulted in synthetic lethality, and we were unable to continue our investigation. The result of synthetic lethality of TLC1 and YKU70 double deletion was shown in the Figure 7B in the reviewed preprint version 1, and the result was not included in the reviewed preprint version 2 in accordance with the reviewer's instructions.
"Atypical” survivors could be detected in SY13 cells (Figure 1D), but the frequency of their formation in the SY13 strain appeared to be lower than in SY12. As one can imagine, SY13 contains two chromosomes and its survivors should have a higher frequency of intra-chromosome fusions.
(2) From their data, it is possible that X and Y elements influence homologous recombination, type 1 and type 2 (type X), at telomeres. In particular, the presence of X and Y elements appears to be important for promoting type 1 recombination. In other words, although not essential, subtelomeres have some function in maintaining telomeres. I suggest that the authors include author response image 4 in the text. They could revise their conclusion and the paper title accordingly.
According to this suggestion, we have included author response image 4 in the revised manuscript as Figure 2E, Figure 5D, Figure 6C and Figure 6E. Accordingly, we have changed the title as “Elimination of subtelomeric repeat sequences exerts little effect on telomere essential functions in Saccharomyces cerevisiae”.
(3) Minor points: The newly added data indicate that X survivors are generated in a type 2-dependent manner. The authors could discuss how Y elements were eroded while retaining X elements (line 225, Figure 2A).
Thank this reviewer’s suggestion. We have discussed it in the revised manuscript (p.13 line 244-245). When telomere was deprotected, chromosome end resection took place. Since SY12 only has one Y’-element, it is hard to search homology sequences to repair the Y’-element in XVI-L. When the X-element in XVI-L was exposed by further resection, it is easier to find homology sequences to repair. So, in Type X survivor the Y’-element was eroded while retaining X-element.
Reviewer #2
I would like to congratulate the authors for their work and the efforts they put in improving the manuscript. The major criticism I had previously, ie testing the genetic requirements for the survivor subtypes, has been met. Below are a few minor comments that don't necessarily require a response.
(1) I think the Author response image 6 could have been included in the manuscript. I understand that the authors don't want to overinterpret survivor subtype frequencies, but this figure would have suggested some implication of Rad51 in the emergence of survivors even in the absence of Y' elements. At this stage, however, it is up to the authors, and leaving this figure out is also fine in my opinion.
According to the suggestion, the author response image 6 has been presented as Figure 6—figure supplement 7.
(2) Chromosome circularization seems to rely on microhomologies. Previously, the authors proposed that SY14 circularization depended on SSA (Wu et al. 2020), but here, since circularization appears to be Rad52-independent, it is likely to be based on MMEJ rather than SSA (although there are contradictory results on Rad52's role in MMEJ in the literature).
Yes, we mentioned it in the revised manuscript.
(3) p. 28 lines 511-513: "The erosion sites and fusion sequences differed from those observed in SY12 tlc1Δ-C1 cells (Figure 2D), suggesting the stochastic nature of chromosomal circularization": I don't think they are necessarily stochastic, because the sequences beyond the telomeres are now modified, the available microhomologies have changed as well.
We agreed with your opinion. In different chromosomes, there tend to be some hotspots for chromosome fusion. For example, in Figure 6C and 6F the resection site in Chr1 and Chr2 was the same in SY12XYΔ+Y tlc1Δ-C1 and SY12XYΔ tlc1Δ-C1. So, we speculate that there are some hotspots for chromosome fusion, but which site the cell will choose in one round chromosome fusion event is stochastic.
(4) Typos and other errors:
- p. 3 line 52: "subtelomerice" and "varies" are mispelled.
- p. 5 line 78: "processes" should be "process".
- Supp files are mislabelled (the numbers do not correspond to file name).
- Supp file 2: how come SY12 has only one Y' element and SY13 has two?
- p. 10 line 175: "emerging" should be "emergence".
- p.15 line 276: "counter-selected" should be "being counter-selected" or "counterselection".
- p. 29 line 523: "the formation of them" should be "their formation".
- p. 37 line 653: "could have been an ideal tool": the sentence is grammatically incorrect. Writing "AND could have been an ideal tool" is enough to make it structurally correct.
Thanks for pointing these errors out. We have corrected them in the revised manuscript. For the question “how come SY12 has only one Y' element and SY13 has two?” we were not sure at this moment. We speculated that one of the Y’ might be lost during genetic engineering of the chromosomes by CRISPR–Cas9 system.
Reviewer #3
The authors included statistical analyses of the qPCR data (Fig 4B) as requested, but did not comment on the striking difference in expression of MPH3 and HSP32 in the SY12 strain compared to BY4742. An improvement of the manuscript is the inclusion of rad52 tlc1 strains in their analyses, demonstrating that the "atypical and circular survivors" arose independently of homologous recombination. In addition, by analyzing rad51 and rad50 mutant strain they could demonstrate that the "type X" survivors had similar molecular requirements to type II survivors. Overall, the revised submission improves the article.
We thank the reviewer’s comments and suggestions. The SY12 strain (with three chromosomes) exhibited lower expression levels of both MPH3 and HSP32 compared to the parental strain BY4742 (with 16 chromosomes). We speculated that with the reduced chromosome numbers, the silencing proteins appeared to no longer be titrated by other telomeres that have been deleted. We have added these comments in the revised manuscript.
Wu, Z.J., Liu, J.C., Man, X., Gu, X., Li, T.Y., Cai, C., He, M.H., Shao, Y., Lu, N., Xue, X., et al. (2020). Cdc13 is predominant over Stn1 and Ten1 in preventing chromosome end fusions. Elife 9.
-

eLife assessment
This important study advances our understanding of the biological significance of the DNA sequence adjacent to telomeres. The data presented convincingly demonstrate that subtelomeric repeats are non-essential and have a minimal, if any, role in maintaining telomere integrity of budding yeast. The work will be of interest to the telomere community specifically and the genome integrity community more broadly.
-

Reviewer #1 (Public Review):
The authors have generated a set of yeast S. cerevisiae strains containing different numbers of chromosomes.
Elimination of telomerase activates homologous recombination (HR) to maintain telomeres in cells containing the original 16 chromosomes. However, elimination of telomerase leads to circularization of cells containing a single or two chromosomes. The authors examined whether the subtelomeric sequences X and Y' promote HR-mediated telomere maintenance using the strain SY12 carrying three chromosomes. They found that the subtelomeric sequences X and Y' are dispensable for cell proliferation and HR-mediated telomere maintenance in telomerase-minus SY12 cells. They conclude that subtelomeric X and Y' sequences do not play essential roles in both telomerase-proficient and telomerase-null cells and propose …Reviewer #1 (Public Review):
The authors have generated a set of yeast S. cerevisiae strains containing different numbers of chromosomes.
Elimination of telomerase activates homologous recombination (HR) to maintain telomeres in cells containing the original 16 chromosomes. However, elimination of telomerase leads to circularization of cells containing a single or two chromosomes. The authors examined whether the subtelomeric sequences X and Y' promote HR-mediated telomere maintenance using the strain SY12 carrying three chromosomes. They found that the subtelomeric sequences X and Y' are dispensable for cell proliferation and HR-mediated telomere maintenance in telomerase-minus SY12 cells. They conclude that subtelomeric X and Y' sequences do not play essential roles in both telomerase-proficient and telomerase-null cells and propose that these sequences represent remnants of genome evolution.Interestingly, telomerase-minus SY12 generates survivors that are different from well-established Type I or Type II survivors. The authors uncover atypical telomere formation which does not depend on the Rad52 homologous recombination pathway.
Strengths:
The authors examined whether the subtelomeric sequences X and Y' promote HR-mediated telomere maintenance using the strain SY12 carrying three chromosomes. They show that subtelomeres do not have essential roles in telomere maintenance and cell proliferation.
Weaknesses:
It is not fully addressed how atypical survivors are generated independently of Rad52-mediated homologous recombination.
It remains possible that X and Y elements influence homologous recombination, type 1 and type 2 (type X), at telomeres. In particular, the presence of X and Y elements appears to be important for promoting type 1 recombination, although the authors conclude "Elimination of subtelomeric repeat sequences exerts little effect on telomere functions". -

Reviewer #2 (Public Review):
Summary:
In this work, Hu and colleagues investigate telomerase-independent survival in Saccharomyces cerevisiae strains engineered to have different chromosome numbers. The authors describe the molecular patterns of survival that change with fewer chromosomes and that differ from the well-described canonical Type I and Type II, including chromosome circularization and other atypical outcomes. They then take advantage of the strain with 3 chromosomes to examine the effect of deleting all the subtelomeric elements, called X and Y'. For most of the tested phenotypes, they find no significant effect of the absence of X- and Y'-element, and show that they are not essential for survivor formation. They speculate that X- and Y'-elements are remnants of ancient telomere maintenance mechanisms.
Strengths:
This work …
Reviewer #2 (Public Review):
Summary:
In this work, Hu and colleagues investigate telomerase-independent survival in Saccharomyces cerevisiae strains engineered to have different chromosome numbers. The authors describe the molecular patterns of survival that change with fewer chromosomes and that differ from the well-described canonical Type I and Type II, including chromosome circularization and other atypical outcomes. They then take advantage of the strain with 3 chromosomes to examine the effect of deleting all the subtelomeric elements, called X and Y'. For most of the tested phenotypes, they find no significant effect of the absence of X- and Y'-element, and show that they are not essential for survivor formation. They speculate that X- and Y'-elements are remnants of ancient telomere maintenance mechanisms.
Strengths:
This work advances our understanding of the telomerase-independent strategies available to the cell by altering the structure of the genome and of the subtelomeres, a feat that was enabled by the set of strains they engineered previously. By using strains with non-standard genome structures, several alternative survival mechanisms are uncovered, revealing the diversity and plasticity of telomere maintenance mechanisms. Overall, the conclusions are well supported by the data, with adequate sample sizes for investigating survivors. The assessment of the genetic requirements for survivors in strains with different chromosome numbers greatly improved the quality of this work. The molecular analyses based on Southern blots are also very well-conducted.
Weaknesses:
The authors discovered alternative telomerase-independent survival strategies beyond the well-described type I and II (including circularization, type X and atypical, as they called them) at play in the context of reduced number of chromosomes. Their work provides a molecular and a partial genetic characterization of these survival pathways. A more thorough analysis of the frequency of each type of survivors and their genetic requirements would have advanced our understanding or the diversity of survival strategies in the absence of telomerase. However, as noted by the authors, the quantification of the rate of emergence of survivors (and their subtypes) is very difficult to achieve. This comment is therefore not meant as a criticism but rather as a perspective on exciting future research avenues.
-

Reviewer #3 (Public Review):
This study investigates subtelomeric repetitive sequences in the budding yeast Saccharomyces cerevisiae, known as Y' and X-elements. Taking advantage of yeast strain SY12 that contains only 3 chromosomes and six telomeres (normal yeast strains contain 32 telomeres) the authors are able to generate a strain completely devoid of Y'- and X-elements.
Strengths:
They demonstrate that the SY12 delta XY strain displays normal growth, with stable telomeres of normal length that were transcriptionally silenced, a key finding with wide implications for telomere biology. Inactivation of telomerase in the SY12 and SY12 delta XY strains frequently resulted in survivors that had circularized all three chromosomes, hence bypassing the need for telomeres altogether. They show that survivors with fused chromosomes and …
Reviewer #3 (Public Review):
This study investigates subtelomeric repetitive sequences in the budding yeast Saccharomyces cerevisiae, known as Y' and X-elements. Taking advantage of yeast strain SY12 that contains only 3 chromosomes and six telomeres (normal yeast strains contain 32 telomeres) the authors are able to generate a strain completely devoid of Y'- and X-elements.
Strengths:
They demonstrate that the SY12 delta XY strain displays normal growth, with stable telomeres of normal length that were transcriptionally silenced, a key finding with wide implications for telomere biology. Inactivation of telomerase in the SY12 and SY12 delta XY strains frequently resulted in survivors that had circularized all three chromosomes, hence bypassing the need for telomeres altogether. They show that survivors with fused chromosomes and so-called atypical survivors arise independently of the central recombination protein Rad52. The SY12 and SY12 delta XY yeast strains can become a useful tool for future studies of telomere biology. The conclusions of this manuscript are well supported by the data and are valuable for researchers studying telomeres.
Weaknesses:
A weakness of the manuscript is the analysis of telomere transcriptional silencing. They state: "The results demonstrated a significant increase in the expression of the MPH3 and HSP32 upon Sir2 deletion, indicating that telomere silencing remains effective in the absence of X and Y'-elements". However, for the SY12 strain, their analyses indicate that the difference between the WT and sir2 strains is nonsignificant. In addition, a striking observation is that the SY12 strain (with only three chromosomes) express much less of both MPH3 and HSP32 than the parental strain BY4742 (16 chromosomes), both in the presence and absence of Sir2.
-

-

Author Response
On behalf of my co-authors, I thank you very much for sending our manuscript (# eLifeRP-RA-2023-91223) entitled “Elimination of subtelomeric repeat sequences exerts little effect on telomere functions in Saccharomyces cerevisiae” for review and providing us an opportunity for revision. We also thank the reviewers for their critical and constructive comments and suggestions which have helped us to strengthen our study. We have performed more experiments to address the concerns the reviewers raised, and we have also revised or corrected some of our statements as the reviewers suggested.
Reviewer #1
- The author’s data indicate that cells with many chromosomes are more dependent on possibly homologous recombination than SY12 cells with three chromosomes. Telomerase-deficient cells exhibit the type I and type II telomere …
Author Response
On behalf of my co-authors, I thank you very much for sending our manuscript (# eLifeRP-RA-2023-91223) entitled “Elimination of subtelomeric repeat sequences exerts little effect on telomere functions in Saccharomyces cerevisiae” for review and providing us an opportunity for revision. We also thank the reviewers for their critical and constructive comments and suggestions which have helped us to strengthen our study. We have performed more experiments to address the concerns the reviewers raised, and we have also revised or corrected some of our statements as the reviewers suggested.
Reviewer #1
- The author’s data indicate that cells with many chromosomes are more dependent on possibly homologous recombination than SY12 cells with three chromosomes. Telomerase-deficient cells exhibit the type I and type II telomere structures, whereas telomerase-deficient SY12 cells often generate different telomere structures (named Type X survivors or atypical survivors). Type I survivor depends on Rad51 possessing tandem Y' elements whereas Type II survivor depends on Rad59 carrying long TG sequences (line 60-70). Both types require Rad52 (line 66-70). At the moment, it is not determined how Type X or atypical survivors are generated in telomerase-deficient SY12 cells.
The authors need to determine whether Type X or atypical survivors depend on other repair pathways from Type I and Type II, and what DNA sequences are retained adjacent to telomeres in Type X or atypical survivors by sequencing analysis (Fig. 2).
We thank the reviewer’s valuable comments and suggestions. Atypical survivor is a subtype of survivor that exhibits non-uniform telomere patterns, distinct from those observed in Type I, Type II, Type X, or circular survivors. To further determine its genetic requirements, we deleted RAD52 in SY12 tlc1Δ, SY12YΔ tlc1Δ, SY12XYΔ tlc1Δ, and SY12XYΔ+Y tlc1Δ strains. Southern blotting results showed that neither Type I nor Type II survivors were found in the series of strains; circular survivor was in the predomination; beside circular survivor, some survivors exhibiting non-uniform telomere patterns suggested they were atypical survivor. These results have been presented as Figure 2—figure supplement 6B, Figure 5—figure supplement 2B and Figure 6—figure supplement 4B in the revised version. The results showed that atypical survivors still emerged when Rad52 pathway was repressed, indicating that the formation of atypical survivors does not strictly rely on the homologous recombination.
Given that "atypical" clones exhibit non-uniform telomere patterns, it’s not surprising that their chromosome structures are variable and tanglesome. Consequently, it is hard for us to amplify and sequence the DNA sequences retained adjacent to telomeres.
Since no Type X survivor was detected in SY12 tlc1Δ rad52Δ strain (Author response image 1A), we deleted RAD50 or RAD51 in SY12 tlc1Δ strain to investigate on which pathway the formation of the Type X survivor relied. Results showed that Type X survivor emerged in the absence of Rad51 but not Rad50, suggesting that the formation of Type X survivor depended on Rad50 pathway. These results have been presented as Figure 2—figure supplement 7.
To determine the chromosomal end structure of the Type X survivor, we randomly selected a typical Type X survivor, and performed PCR-sequencing analysis. The results revealed the intact chromosome ends for I-L, X-R, XIII-L, XI-R, and XIV-R, albeit with some mismatches compared with the S. cerevisiae S288C genome, which possibly arising from recombination events that occurred during survivor formation. Notably, the sequence of the Y’-element in XVI-L could not be detected, while the X-element remained intact. Figure 2—figure supplement 5 in the revised manuscript.
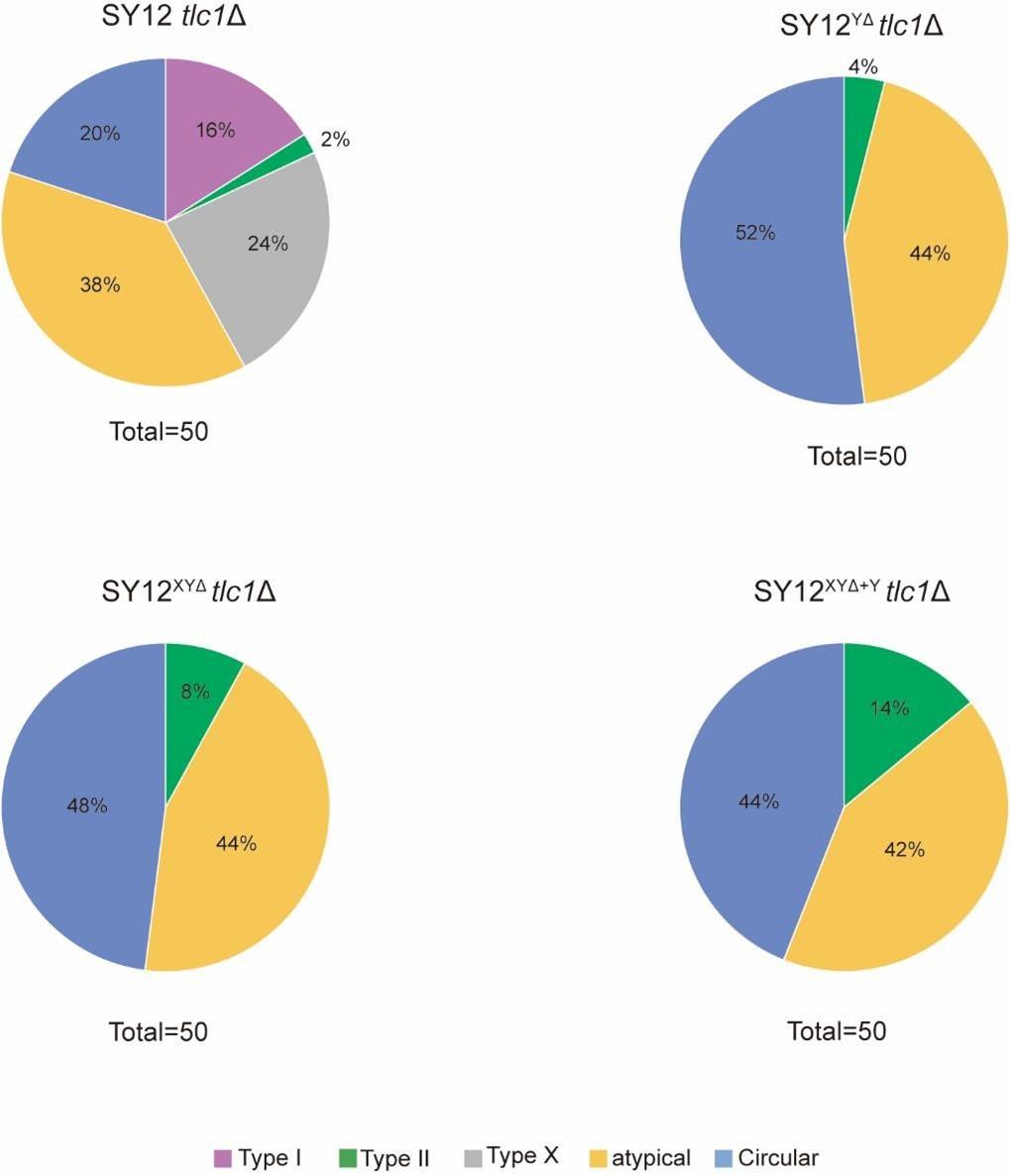
- Survivor generation of each type (Type I, Type II, Type X or atypical and circularization) needs to be accurately quantitated. The authors concluded that X or Y' elements are not strictly necessary for survivor formation (Fig. 5 and Fig. 6). However, their removal appears to increase atypical survivor and chromosome circularization (Fig. 2 vs Fig. 5 and 6).
We are grateful for the reviewer’s critical and constructive suggestions. According to the reviewer’s requirement, we quantified each type of survivors in SY12 tlc1Δ, SY12YΔ tlc1Δ, SY12XYΔ tlc1Δ and SY12XYΔ+Y tlc1Δ strains (Figure 2D, 5C, 6A and 6B). In SY12 tlc1Δ strain, Type I survivors accounted for 16%, Type II survivors for 2%, Type X survivors for 24%, circular survivors for 20% and atypical survivors for 38%. In SY12YΔ tlc1Δ strain, 4% were Type II survivors, 52% were circular survivors and 44% were atypical survivors.
For the SY12XYΔ tlc1Δ strain, 8% were Type II survivors, 48% were circular survivors and 44% were atypical survivors. In SY12XYΔ+Y tlc1Δ strain, the proportions of Type II, circular and atypical survivors were 14%, 44%, and 42%, respectively (Author response image 1).
In comparing SY12YΔ with SY12XYΔ, we observed a similar ratio of circular and atypical survivors. This result indicates that the remove of X-elements exert little effect on the formation of circular and atypical survivors. Similarly, in SY12XYΔ+Y strain, the proportions of circular and atypical survivors were comparable to those in SY12XYΔ strain, indicating that Y’-elements also have little effect on the formation of circular and atypical survivors. However, due to the unknown frequency of survivor formation, alternative explanations of these data are possible. For example, subtelomeric elements previously suggested to have no impact on the formation of any survivor types might influence every type to similar extents, leading to similar ratios across all survivor types. With our present data, it is still unclear whether the absence of X and Y'-elements enhances the formation of circular and atypical survivors. Therefore, we did not present these results in the revised manuscript.
Author response image 1.
Quantitation of each survivor type in SY12 subtelomerice engineered strains. The ratio of survivor types in SY12 tlc1Δ, SY12YΔ tlc1Δ, SY12XYΔ tlc1Δ and SY12XYΔ+Y tlc1Δ strains. Type I, pulper; Type II, green; Type X, gray; atypical survivor, orange; circular survivor, blue.
3)The authors asked whether X and Y' elements are required for cell proliferation, stress response, telomere length control and telomere silencing (Fig. 4). Similar studies have been previously carried out by using synthetic chromosomes (see PMID: 28300123). The authors need to discuss this point.
Thanks for your suggestion, we have added the information in the revised version. (p.24 line 449-453)
- The Fig. 7 data support that circular chromosomes do not require Ku-dependent DNA end protection. This is consistent with the current view that Ku binds and protects DNA ends. This finding by itself does not contribute significantly to our understanding of telomere maintenance. The authors need to more extensively discuss the significance of their findings in SY12 cells compared to wild-type cells with 16 chromosomes.
We agree with the logic that this reviewer has pointed out. Our results demonstrate that combinatorial deletion of YKU70 and TLC1 caused synthetic lethality in SY12 cells, which possess three linear chromosomes, However, it did not affect the viability of "circular survivors", supporting the notion that telomere deprotection leads to the synthetic lethality in yku70Δ tlc1Δ double mutants. Nevertheless, this conclusion merely confirms the current view observed in wild-type cells that Ku binds and protects DNA ends.
To avoid confusing readers and maintain the logical flow of the manuscript, we have deleted this section in the revised version.
Minor issues:
- Line 112-113: " for SY13, which contains two chromosomes, could also have a high probability of circularizing all chromosomes for survival": The reference or the supplemental data are required.
Thank this reviewer for the suggestion. According to the reviewer’s comments, we performed a Southern blotting assay to examine the types of survivors in SY13 tlc1Δ strain. We found that the majority of SY13 tlc1Δ clones exhibited hybridization signal similar to SY14 tlc1Δ circular survivors, pointing to the possibility that two chromosomes in these survivors may undergo intra-chromosomal fusions. This result has been added to figure 1D in the revised version.
- Line 349-350: The BY4742 mre11Δ haploid strain serves as a negative control. The authors need to explain why mre11 cells serve as a negative control.
Thank this reviewer for the comment. We employed mre11Δ as negative control because Mre11 is a member of the RAD52 epistasis group, which is involved in the repair of double-stranded breaks in DNA, and mutants in MRE11 exhibit defects in the repair of DNA damages caused by DNA damage drugs (Krogh and Symington, 2004; Lewis et al., 2004; Symington, 2002). (p.23 line 420-422)
Reviewer #2
- The qualification of survivor types mostly relies on molecular patterns in Southern blots. While this is a valid method for a standard strain, it might be more difficult to apply to the strains used in this study. For example, in SY8, SY11 and SY12, the telomere signal at 1-1.2 kb can be very faint due to the small number of terminal Y' elements left. As another example, for the Y'-less strain, it might seem obvious that no Type I survivor can emerge given that Y' amplification is a signature of Type I, but maybe Type-I-specific molecular mechanisms might still be used. To reinforce the characterization of survivor types, an analysis of the genetic requirements for Type I and Type II survivors (e.g. RAD51, RAD54, RAD59, RAD50) could complement the molecular characterization in specific result sections.
We thank this reviewer for his/her constructive comments and suggestions. To investigate whether Type-I-specific molecular mechanisms are still utilized in the survivor formation in Y'-less strain, we deleted RAD51 in SY12XYΔ tlc1Δ. SY12XYΔ tlc1Δ rad51Δ strain was able to generate three types of survivors, including Type II survivor, circular survivor and atypical survivor, similar to the observations in SY12XYΔ tlc1Δ strain. However, the ratios of circular and atypical survivors were 36% and 32%, respectively, lower than the 48% and 44% observed in SY12XYΔ tlc1Δ strain (supplementary file 5). This result indicates that Type-I-specific molecular mechanisms contribute to the survivor formation. Given that our work primarily focuses on the function of subtelomeric elements, we chose not to include this result in our revised manuscript to maintain a coherent logical flow.
To reinforce the characterization of survivor types, we deleted RAD50, RAD51 and RAD52 in SY12 tlc1Δ strain, respectively. Southern blotting assay revealed that in the absence of Rad51, no Type I survivor was detected; in the absence of Rad50, neither Type I nor Type X survivor was detected. However, circular and atypical survivors still emerged in the absence of Rad52, suggesting that the RAD52-mediated homologous recombination is not strictly necessary for the formation of circular and atypical survivors. These results have been presented as Figure 2—figure supplement 6 and Figure 2— figure supplement 7.
- In the title, the abstract and throughout the discussion, the authors chose to focus on the effect of X- and Y'-element deletion on different phenotypes and on survivor formation, as the main message to convey. While it is a legitimate and interesting message, other important results of this work might benefit from more spotlight. Namely, the observation that strains with different chromosome numbers show different survivor patterns and that several survival strategies beyond Type I and II exist and can reach substantial frequencies depending on the chromosomal context.
Thanks for your valuable suggestion. While we value your suggestion to highlight additional aspects of our work, we would like to express our perspective on the current emphasis on the effect of X- and Y'-element deletion. We believe that by maintaining this focus, we can present a more coherent and impactful narrative for our readers. Additionally, we recognize that the relationship between chromosome numbers and survivor type frequencies is complex and warrants further experimental validation. We are considering exploring this aspect in more detail in our future projects. However, we fully acknowledge the importance of the observations you raised concerning strains with different chromosome numbers and the diversity of survival strategies.
- In SY12 strain, while X- and Y'-elements are not essential for survivor emergence, they do modulate the frequency of each type of survivors, with more chromosome circularization events observed for SY12YΔ, SY12XYΔ and SY12XYΔ+Y strains. This result should be stated and discussed, maybe alongside the change in survivor patterns in the other SY strains, to more accurately assess the roles of these subtelomeric elements.
Following the reviewer’s suggestion, we compared the circular survivor ratios in SY12 tlc1Δ, SY12YΔ tlc1Δ, SY12XYΔ tlc1Δ and SY12XYΔ+Y tlc1Δ strains (supplementary file 5). It appears that the formation of circular survivors is less efficient in the SY12 tlc1Δ, with a ratio of 20%, much lower than that in SY12YΔ tlc1Δ, SY12XYΔ tlc1Δ or SY12XYΔ+Y tlc1Δ strains. However, it should be noted that SY12 tlc1Δ can generate Type I and Type X survivors, potentially decreasing the ratio of circular survivors.
Therefore, we further compared the circular survivor ratios in SY12YΔ tlc1Δ, SY12XYΔ tlc1Δ and SY12XYΔ+Y tlc1Δ strains. In the SY12YΔ tlc1Δ strain, circular survivors accounted for 52% (26/50), comparable to 48% (24/50) in the SY12XYΔ tlc1Δ strain, indicating that X- elements exert little effect on the formation of circular survivor. Additionally, the ratio of circular survivors was 44% (22/50) in SY12XYΔ+Y tlc1Δ strain, also comparable to 48% (24/50) in the SY12XYΔ tlc1Δ strain, suggesting that Y’-element also has little effect on chromosome circularization. However, due to the unknown frequency of survivor formation, alternative explanations of these data are possible. For example, subtelomeric elements previously suggested to have no impact on the formation of any survivor types might influence every type to similar extents, resulting in similar ratios across all survivor types. With our current data, it is still uncertain whether X and Y'-elements modulate the frequency of each type of survivors. Therefore, we did not include these results in the revised manuscript.
- The authors might want to update some general information about subtelomere structure and their diversity across yeast strain with the recent paper by O'Donnell et al. 2023 Nature Genetics, "Telomere-to-telomere assemblies of 142 strains characterize the genome structural landscape in Saccharomyces cerevisiae".
Thanks for your advice. We have added this information in the revised manuscript. (p.3 line 51-54)
- Although it is cited in the discussion, the recent work by the Malkova lab (Kockler et al. 2021 Mol Cell) could be mentioned in the introduction as it conceptually changes our views on survivor formation, its dynamics and the categorization into Type I and Type II.
Thanks for your advice. We have added this information in the revised manuscript. (p.5 line 75-78)
- p.7 line 128-130: rather than chromosome number, the ratio of survivor types might be controlled by the fraction of subtelomeres with Y'-elements and their relative configuration across chromosomes. A map of the structure of remaining subtelomeres in the SYn strains might be good to have.
We have added this information in supplementary file 2 in the revised manuscript.
- Fig. 1C: in SY9 tlc1Δ, the lane with triangle mark looks like a type II.
The hybridization pattern of SY9 tlc1Δ clone 2 has both amplified Y’L-element and long heterogeneous TG1-3 repeats, it might be the “hybrid” survivor mentioned by Kockler’s work (Kockler et al., 2021). Therefore, we classify it as a no-classical survivor.
- p.9 line 149: the title of this result section "Y'-element is not essential for the viability of cells carrying linear chromosomes" doesn't reflect well the content of the section, which is more about characterizing the survivor pattern in SY12.
Thanks for your advice. We have changed the title of this section into “Characterizing the survivor pattern in SY12” in the revised manuscript. (p.9 line 155)
- p.10 line 167: that type I can emerge in SY12 indicates that multiple Y'-elements in tandem are not required for type I recombination. I am not sure if this was already known, but it could be noted.
We appreciate the reviewer’s comment. We have added this information in the revised manuscript. (p.10 175-177)
- p.18 line 318-320: the deletion of the Y' element also seems to remove the centromere-proximal telomere sequence adjacent to it. Maybe it should be stated as well. Even more importantly, in lines 327-329, the Y'-element that is reintroduced in the strain does not include the centromere-proximal short telomere sequence. This is important to interpret the Southern blots.
We thank the reviewer for this critical suggestion. The deletion of Y'-element including both Y’- and X- element sequence in XVI-L (supplementary file 4), and the Y’element in the XVI-L does not contain the centromere-proximal telomere sequence. The Y'-element reintroduced into the left arm of Chr 3 in SY12XYΔ strain was cloned from native left arm of XVI in SY12 strain which does not contain the centromere-proximal short telomere sequence. Besides listing these details in supplementary file 4, we also emphasize it in the revised manuscript (p.21 line 397-398).
- p.29 lines 496-497: it seems that X and Y'-elements tend to inhibit formation of circular survivors either directly (by participating in end protection), or by promoting type I and type II, thus reducing the fraction of circular survivors. Maybe this could be added to the conclusion of this section.
We thank the reviewer for his/her comments and have analyzed survivor types in SY12 tlc1Δ, SY12YΔ tlc1Δ, SY12XYΔ tlc1Δ and SY12XYΔ+Y tlc1Δ strains (supplementary file 5). Circular survivor formation appears less efficient in the SY12 tlc1Δ, with a ratio of 20%, significantly lower than SY12YΔ tlc1Δ, SY12XYΔ tlc1Δ or SY12XYΔ+Y tlc1Δ strains. However, it is noteworthy that SY12 tlc1Δ can generate Type I and Type X survivors, potentially impacting the circular survivor ratio.
We further compared circular survivor ratios in SY12YΔ tlc1Δ, SY12XYΔ tlc1Δ and SY12XYΔ+Y tlc1Δ strains. SY12YΔ tlc1Δ had 52% circular survivors, similar to SY12XYΔ tlc1Δ with 48%, indicating minimal impact of X- elements. Additionally, SY12XYΔ+Y tlc1Δ had 44% circular survivors, also similar to SY12XYΔ tlc1Δ, suggesting that Y’-element has little effect on chromosome circularization. However, due to unknown frequency of survivor formation, alternative explanations, like subtelomeric elements affecting all the type of survivor similarly, are possible. With our current data, it remains unclear whether X and Y'-elements are involved in end protection and consequently inhibit the formation of circular survivors.
Therefore, these results were not included in the revised manuscript.
- p.32 line 533: this result section doesn't really fit with the rest of the paper, does it?
Thanks for your valuable advice. To avoid confusing readers and to keep the fluency of logic flow of the manuscript we have deleted this section in the revised version.
- The methods section does not describe the experiments sufficiently and it often lacks specific details such as the manufacturer or references. Some sections of the methods are more exhaustive than others. They should all be written with the same level of detail in my opinion.
Thanks for your advice. We have described the experiments more sufficiently and added the manufacturer or references in the ‘materials and methods’ part in the revised manuscript. (p.41 line741-745, p.42 line 755-756, p.42 line 762-770, p.43 line 788 and p.45 line 812-813)
Minor comments, typos and grammatical errors:
p.3 line 33: "INTROUDUCTION" should be "INTRODUCTION".
We have corrected it in the revised manuscript. (p.3 line 33) p.4 line 54: "S, cerevisiae", use dot instead of comma. R15: We have corrected it in the revised manuscript. (p.4 line 57)
p.4 line 55: I believe TLC1 as the RNA moiety should be in (non-italicized) capital letters and not written as a protein.
We have corrected it in the revised manuscript. (p.4 line 58)
p.7 line 115: please indicate that pRS316 uses URA3 as a marker, otherwise the counterselection with 5'-FOA is not obvious.
Thank this reviewer for the comment. We have added this statement in the revised manuscript. (p.7 line 121-122)
p.12 line 206: tlc1Δ should be in italic.
We have corrected it in the revised manuscript. (p.10 line 184)
p.13 lines 227-229: "where only one hybridization signal", a verb seems to be missing.
We thank the reviewer’s kind reminder and have corrected the mentioned errors in the revised manuscript. (p.14 line 254-255)
Reviewer #3
- A weakness of the manuscript is the analysis of telomere transcriptional silencing. They state: "The results demonstrated a significant increase in the expression of the MPH3 and HSP32 upon Sir2 deletion, indicating that telomere silencing remains effective in the absence of X and Y'-elements". However, there are no statistical analyses performed as far as I can see. For some of the strains, the significance of the increased expression in sir2 (especially for MPH3) looks questionable. In addition, a striking observation is that the SY12 strain (with only three chromosomes) express much less of both MPH3 and HSP32 than the parental strain BY4742 (16 chromosomes), both in the presence and absence of Sir2. In fact, the expression of both MPH3 and HSP32 in the SY12 sir2 strain is lower than in the BY4742 SIR2+ strain. In addition, relating this work to previous studies of subtelomeric sequences in other organisms would make the discussion more interesting.
First, I enjoyed reading your manuscript. It would be great if you performed the statistical analysis on the RT-qPCR data in figure 4B and addressed the issue of the difference of the BY4742 and SY12 strains. A model could be that this is a titration effect of silencing proteins due to fewer telomeres, which could be investigated by performing the analyses on more SY-strains with variable numbers of telomeres.
We highly appreciate the reviewer’s valuable comments and suggestions, which included a point that has also left us confused. We conducted statistical analyses on the RT-qPCR data, and the t-test result revealed that upon the deletion of Sir2, SY12YΔ, SY12XYΔ and SY12XYΔ+Y strains exhibited a significant increase in MPH3 expression (located on the right arm of chr X) with a P value < 0.05. In the case of SY12, the deletion of Sir2 resulted in an increase in gene expression (P value < 0.1). Similar tendencies were observed in the BY4742 strain. The statistical analyses of RTqPCR results on XVI-L mirrored those of X-R.
The results demonstrated a significant increase in MPH3 and HSP32 expression upon SIR2 deletion in SY12YΔ, SY12XYΔ and SY12XYΔ+Y strains, leading to the conclusion that telomere silencing remains effective in the absence of X-and Y’-elements. However, as the reviewer has pointed out, no statistically significant differences in MPH3 and HSP32 expression were observed between the SY12 and SY12 sir2Δ strain. For HSP32, this lack of significance may be attributed to the greater distance between HSP32 and telomere XVI-L in SY12 compared to SY12YΔ, SY12XYΔ or SY12XYΔ+Y strains, resulting in a weaker telomere position effect on HSP32 and a non-significant increase in gene expression in SY12. However, this explanation does not apply to MPH3, as SY12YΔ, with a same distance between MPH3 and telomere X-R as in SY12, still exhibits an effective telomere position effect on MPH3. We cannot provide a compelling explanation at this moment, and we suspect that the lack of statistically significant differences may be due to random clonal variation.
Additionally, the SY12 strain (with three chromosomes) exhibited lower expression levels of both MPH3 and HSP32 compared to the parental strain BY4742 (with 16 chromosomes). Notably, it has been reported that the expression of genes coding silencing proteins in SY14 (with one chromosomes) were nearly identical to that of BY4742 (with 16 chromosomes)(Shao et al., 2018). Consequently, with respect to the reduced chromosome numbers, the silencing proteins appeared to be relatively overexpressed. Therefore, as pointed out by the reviewer, this observed phenomenon may be attributed to a titration effect of silencing proteins due to fewer telomeres. We have added the statistical analyses result in Figure 4B.
We have related our work with previous studies of subtelomeric sequences in fission yeast in the discussion part. (p.37 line 655-676)
Minor points are to correct the figure legend for Figure 6 supplement 1 (the strain designations) and line 55, RNAs are written with all caps, i.e. TLC1, and line 537 delete the "which" in the sentence.
Thanks for your advice. We have corrected them in the revised manuscript.
The strain has been replaced with SY12XYΔ+Y (p.35 line 617, 618 and 620)
“Tlc1” has been replaced with “TLC1” (p.4 line 58).
We have deleted the section of “Circular chromosome maintain stable when double knockout of yku70 and tlc1” according to the suggestions raised by reviewer 1 and 2, the deleted section contain the sentence in line 537 you mentioned.
Kockler, Z.W., Comeron, J.M., and Malkova, A. (2021). A unified alternative telomerelengthening pathway in yeast survivor cells. Molecular Cell 81, 1816-1829.e1815. Krogh, B.O., and Symington, L.S. (2004). Recombination proteins in yeast. Annu Rev Genet 38, 233-271.
Lewis, L.K., Storici, F., Van Komen, S., Calero, S., Sung, P., and Resnick, M.A. (2004). Role of the nuclease activity of Saccharomyces cerevisiae Mre11 in repair of DNA double-strand breaks in mitotic cells. Genetics 166, 1701-1713.
Shao, Y., Lu, N., Wu, Z., Cai, C., Wang, S., Zhang, L.L., Zhou, F., Xiao, S., Liu, L., Zeng, X., et al. (2018). Creating a functional single-chromosome yeast. Nature 560, 331-335. Symington, L.S. (2002). Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev 66, 630-670, table of contents.
-

eLife assessment
This important study advances our understanding of the biological significance of the DNA sequence adjacent to telomeres. The data presented convincingly demonstrates that subtelomeric repeats are non-essential and have a minimal, if any, role in maintaining telomere integrity of budding yeast. The work will be of interest to telomere community specifically and the genome integrity community more broadly.
-

Reviewer #1 (Public Review):
The authors have generated a set of yeast S. cerevisiae strains containing different numbers of chromosomes.
Elimination of telomerase activates homologous recombination (HR) to maintain telomeres in cells containing the original 16 chromosomes. However, elimination of telomerase leads to circularization of cells containing a single or two chromosomes. The authors examined whether the subtelomeric sequences X and Y' promote HR-mediated telomere maintenance using the strain SY12 carrying three chromosomes. They found that the subtelomeric sequences X and Y' are dispensable for cell proliferation and HR-mediated telomere maintenance in telomerase-minus SY12 cells. They conclude that subtelomeric X and Y' sequences do not play essential roles in both telomerase-proficient and telomerase-null cells and propose …Reviewer #1 (Public Review):
The authors have generated a set of yeast S. cerevisiae strains containing different numbers of chromosomes.
Elimination of telomerase activates homologous recombination (HR) to maintain telomeres in cells containing the original 16 chromosomes. However, elimination of telomerase leads to circularization of cells containing a single or two chromosomes. The authors examined whether the subtelomeric sequences X and Y' promote HR-mediated telomere maintenance using the strain SY12 carrying three chromosomes. They found that the subtelomeric sequences X and Y' are dispensable for cell proliferation and HR-mediated telomere maintenance in telomerase-minus SY12 cells. They conclude that subtelomeric X and Y' sequences do not play essential roles in both telomerase-proficient and telomerase-null cells and propose that these sequences represent remnants of genome evolution.
Interestingly, telomerase-minus SY12 generate survivors that are different from well-established Type I or Type II survivors. The authors uncover atypical telomere formation which does not depend on the Rad52 homologous recombination pathway.Strengths: The authors examined whether the subtelomeric sequences X and Y' promote HR-mediated telomere maintenance using the strain SY12 carrying three chromosomes. They show that subtelomeres do not have essential roles in telomere maintenance and cell proliferation.
Weaknesses:
It is not fully addressed how atypical survivors are generated independently of Rad52-mediated homologous recombination.
It remains possible that X and Y elements influence homologous recombination, type 1 and type 2 (type X), at telomeres. In particular, the presence of X and Y elements appears to be important for promoting type 1 recombination, although the authors conclude "Elimination of subtelomeric repeat sequences exerts little effect on telomere functions". -

Reviewer #2 (Public Review):
Summary:
In this work, Hu and colleagues investigate telomerase-independent survival in Saccharomyces cerevisiae strains engineered to have different chromosome numbers. The authors describe the molecular patterns of survival that change with fewer chromosomes and that differ from the well-described canonical Type I and Type II, including chromosome circularization and other atypical outcomes. They then take advantage of the strain with 3 chromosomes to examine the effect of deleting all the subtelomeric elements, called X and Y'. For most of the tested phenotypes, they find no significant effect of the absence of X- and Y'-element, and show that they are not essential for survivor formation. They speculate that X- and Y'-elements are remnants of ancient telomere maintenance mechanisms.Strengths:
This work …Reviewer #2 (Public Review):
Summary:
In this work, Hu and colleagues investigate telomerase-independent survival in Saccharomyces cerevisiae strains engineered to have different chromosome numbers. The authors describe the molecular patterns of survival that change with fewer chromosomes and that differ from the well-described canonical Type I and Type II, including chromosome circularization and other atypical outcomes. They then take advantage of the strain with 3 chromosomes to examine the effect of deleting all the subtelomeric elements, called X and Y'. For most of the tested phenotypes, they find no significant effect of the absence of X- and Y'-element, and show that they are not essential for survivor formation. They speculate that X- and Y'-elements are remnants of ancient telomere maintenance mechanisms.Strengths:
This work advances our understanding of the telomerase-independent strategies available to the cell by altering the structure of the genome and of the subtelomeres, a feat that was enabled by the set of strains they engineered previously. By using strains with non-standard genome structures, several alternative survival mechanisms are uncovered, revealing the diversity and plasticity of telomere maintenance mechanisms. Overall, the conclusions are well supported by the data, with adequate sample sizes for investigating survivors. The assessment of the genetic requirements for survivors in strains with different chromosome numbers greatly improved the quality of this work. The molecular analyses based on Southern blots are also very well-conducted.Weaknesses:
The authors discovered alternative telomerase-independent survival strategies beyond the well-described type I and II (including circularization, type X and atypical, as they called them) at play in the context of reduced number of chromosomes. Their work provides a molecular and a partial genetic characterization of these survival pathways. A more thorough analysis of the frequency of each type of survivors and their genetic requirements would have advanced our understanding or the diversity of survival strategies in the absence of telomerase. However, as noted by the authors, the quantification of the rate of emergence of survivors (and their subtypes) is very difficult to achieve. This comment is therefore not meant as a criticism but rather as a perspective on exciting future research avenues. -

Reviewer #3 (Public Review):
This study investigates subtelomeric repetitive sequences in the budding yeast Saccharomyces cerevisiae, known as Y' and X-elements. Taking advantage of yeast strain SY12 that contains only 3 chromosomes and six telomeres (normal yeast strains contain 32 telomeres) the authors are able to generate a strain completely devoid of Y'- and X-elements.
Strengths: They demonstrate that the SY12 delta XY strain displays normal growth, with stable telomeres of normal length that were transcriptionally silenced, a key finding with wide implications for telomere biology. Inactivation of telomerase in the SY12 and SY12 delta XY strains frequently resulted in survivors that had circularized all three chromosomes, hence bypassing the need for telomeres altogether. They show that survivors with fused chromosomes and …
Reviewer #3 (Public Review):
This study investigates subtelomeric repetitive sequences in the budding yeast Saccharomyces cerevisiae, known as Y' and X-elements. Taking advantage of yeast strain SY12 that contains only 3 chromosomes and six telomeres (normal yeast strains contain 32 telomeres) the authors are able to generate a strain completely devoid of Y'- and X-elements.
Strengths: They demonstrate that the SY12 delta XY strain displays normal growth, with stable telomeres of normal length that were transcriptionally silenced, a key finding with wide implications for telomere biology. Inactivation of telomerase in the SY12 and SY12 delta XY strains frequently resulted in survivors that had circularized all three chromosomes, hence bypassing the need for telomeres altogether. They show that survivors with fused chromosomes and so-called atypical survivors arise independently of the central recombination protein Rad52. The SY12 and SY12 delta XY yeast strains can become a useful tool for future studies of telomere biology. The conclusions of this manuscript are well supported by the data and are valuable for researchers studying telomeres.
Weaknesses: A weakness of the manuscript is the analysis of telomere transcriptional silencing. They state: "The results demonstrated a significant increase in the expression of the MPH3 and HSP32 upon Sir2 deletion, indicating that telomere silencing remains effective in the absence of X and Y'-elements". However, for the SY12 strain, their analyses indicate that the difference between the WT and sir2 strains is nonsignificant. In addition, a striking observation is that the SY12 strain (with only three chromosomes) express much less of both MPH3 and HSP32 than the parental strain BY4742 (16 chromosomes), both in the presence and absence of Sir2.
-

-

eLife assessment
This important study advances our understanding of the biological significance of the DNA sequence adjacent to telomeres. The data presented convincingly demonstrates that subtelomeric repeats are non-essential and have a minimal, if any, role in maintaining telomere integrity of budding yeast. The work will be of interest to telomere community specifically and the genome integrity community more broadly.
-

Reviewer #1 (Public Review):
The authors have generated a set of yeast S. cerevisiae strains containing different numbers of chromosomes. Elimination of telomerase activates homologous recombination (HR) to maintain telomeres in cells containing the original 16 chromosomes. However, elimination of telomerase leads to circularization of cells containing a single or two chromosomes. The authors examined whether the subtelomeric sequences X and Y' promote HR-mediated telomere maintenance using the strain SY12 carrying three chromosomes. They found that the subtelomeric sequences X and Y' are dispensable for cell proliferation and HR-mediated telomere maintenance in telomerase-minus SY12 cells. They conclude that subtelomeric X and Y' sequences do not play essential roles in both telomerase-proficient and telomerase-null cells and propose …
Reviewer #1 (Public Review):
The authors have generated a set of yeast S. cerevisiae strains containing different numbers of chromosomes. Elimination of telomerase activates homologous recombination (HR) to maintain telomeres in cells containing the original 16 chromosomes. However, elimination of telomerase leads to circularization of cells containing a single or two chromosomes. The authors examined whether the subtelomeric sequences X and Y' promote HR-mediated telomere maintenance using the strain SY12 carrying three chromosomes. They found that the subtelomeric sequences X and Y' are dispensable for cell proliferation and HR-mediated telomere maintenance in telomerase-minus SY12 cells. They conclude that subtelomeric X and Y' sequences do not play essential roles in both telomerase-proficient and telomerase-null cells and propose that these sequences represent remnants of genome evolution.
Interestingly, telomerase-minus SY12 generate survivors that are different from Type I or Type II survivors.Strengths: The authors examined whether the subtelomeric sequences X and Y' promote HR-mediated telomere maintenance using the strain SY12 carrying three chromosomes.
Weaknesses:
It is not determined how atypical survivors or Type X survivors are generated in telomerase-deficient SY12 cells.
Survivor generation of each type (Type I, Type II, Type X or atypical and circularization) is not quantitated. -

Reviewer #2 (Public Review):
Summary:
In this work, Hu and colleagues investigate telomerase-independent survival in Saccharomyces cerevisiae strains engineered to have different chromosome numbers. The authors describe the molecular patterns of survival that change with fewer chromosomes and that differ from the well-described canonical Type I and Type II, including chromosome circularization and other atypical outcomes. They then take advantage of the strain with 3 chromosomes to examine the effect of deleting all the subtelomeric elements, called X and Y'. For most of the tested phenotypes, they find no significant effect of the absence of X- and Y'-element, and show that they are not essential for survivor formation. They speculate that X- and Y'-elements are remnants of ancient telomere maintenance mechanisms.Strengths:
This work …Reviewer #2 (Public Review):
Summary:
In this work, Hu and colleagues investigate telomerase-independent survival in Saccharomyces cerevisiae strains engineered to have different chromosome numbers. The authors describe the molecular patterns of survival that change with fewer chromosomes and that differ from the well-described canonical Type I and Type II, including chromosome circularization and other atypical outcomes. They then take advantage of the strain with 3 chromosomes to examine the effect of deleting all the subtelomeric elements, called X and Y'. For most of the tested phenotypes, they find no significant effect of the absence of X- and Y'-element, and show that they are not essential for survivor formation. They speculate that X- and Y'-elements are remnants of ancient telomere maintenance mechanisms.Strengths:
This work advances our understanding of the telomerase-independent strategies available to the cell by altering the structure of the genome and of the subtelomeres, a feat that was enabled by the set of strains they engineered previously. By using strains with non-standard genome structures, several alternative survival mechanisms are uncovered, revealing the diversity and plasticity of telomere maintenance mechanisms. Overall, the conclusions are well supported by the data, with adequate sample sizes for investigating survivors. The molecular analyses mostly based on Southern blots are also very well-conducted.Weaknesses:
The qualification of survivor types mostly relies on molecular patterns in Southern blots. While this is a valid method for a standard strain, it might be more difficult to apply to the strains used in this study. For example, in SY8, SY11 and SY12, the telomere signal at 1-1.2 kb can be very faint due to the small number of terminal Y' elements left. As another example, for the Y'-less strain, it might seem obvious that no Type I survivor can emerge given that Y' amplification is a signature of Type I, but maybe Type-I-specific molecular mechanisms might still be used. To reinforce the characterization of survivor types, an analysis of the genetic requirements for Type I and Type II survivors (e.g. RAD51, RAD54, RAD59, RAD50) could complement the molecular characterization in specific result sections.In the title, the abstract and throughout the discussion, the authors chose to focus on the effect of X- and Y'-element deletion on different phenotypes and on survivor formation, as the main message to convey. While it is a legitimate and interesting message, other important results of this work might benefit from more spotlight. Namely, the observation that strains with different chromosome numbers show different survivor patterns and that several survival strategies beyond Type I and II exist and can reach substantial frequencies depending on the chromosomal context.
In SY12 strain, while X- and Y'-elements are not essential for survivor emergence, they do modulate the frequency of each type of survivors, with more chromosome circularization events observed for SY12Y∆, SY12XY∆ and SY12XY∆+Y strains. This result should be stated and discussed, maybe alongside the change in survivor patterns in the other SY strains, to more accurately assess the roles of these subtelomeric elements.
-

Reviewer #3 (Public Review):
Summary:
This study investigates subtelomeric repetitive sequences in the budding yeast Saccharomyces cerevisiae, known as Y' and X-elements. Taking advantage of yeast strain SY12 that contains only 3 chromosomes and six telomeres (normal yeast strains contain 32 telomeres) the authors are able to generate a strain completely devoid of Y'- and X-elements.Strengths: They demonstrate that the SY12 delta XY strain displays normal growth, with stable telomeres of normal length that were transcriptionally silenced, a key finding with wide implications for telomere biology. Inactivation of telomerase in the SY12 and SY12 delta XY strains frequently resulted in survivors that had circularized all three chromosomes, hence bypassing the need for telomeres altogether. The SY12 and SY12 delta XY yeast strains can …
Reviewer #3 (Public Review):
Summary:
This study investigates subtelomeric repetitive sequences in the budding yeast Saccharomyces cerevisiae, known as Y' and X-elements. Taking advantage of yeast strain SY12 that contains only 3 chromosomes and six telomeres (normal yeast strains contain 32 telomeres) the authors are able to generate a strain completely devoid of Y'- and X-elements.Strengths: They demonstrate that the SY12 delta XY strain displays normal growth, with stable telomeres of normal length that were transcriptionally silenced, a key finding with wide implications for telomere biology. Inactivation of telomerase in the SY12 and SY12 delta XY strains frequently resulted in survivors that had circularized all three chromosomes, hence bypassing the need for telomeres altogether. The SY12 and SY12 delta XY yeast strains can become a useful tool for future studies of telomere biology. The conclusions of this manuscript are mostly well supported by the data and are important for researchers studying telomeres.
Weaknesses: A weakness of the manuscript is the analysis of telomere transcriptional silencing. They state: "The results demonstrated a significant increase in the expression of the MPH3 and HSP32 upon Sir2 deletion, indicating that telomere silencing remains effective in the absence of X and Y'-elements". However, there are no statistical analyses performed as far as I can see. For some of the strains, the significance of the increased expression in sir2 (especially for MPH3) looks questionable. In addition, a striking observation is that the SY12 strain (with only three chromosomes) express much less of both MPH3 and HSP32 than the parental strain BY4742 (16 chromosomes), both in the presence and absence of Sir2. In fact, the expression of both MPH3 and HSP32 in the SY12 sir2 strain is lower than in the BY4742 SIR2+ strain. In addition, relating this work to previous studies of subtelomeric sequences in other organisms would make the discussion more interesting.
-