The zinc-finger transcription factor Sfp1 imprints specific classes of mRNAs and links their synthesis to cytoplasmic decay
Curation statements for this article:-
Curated by eLife
eLife assessment
This important study reports that a transcription factor stimulating mRNA synthesis can stabilize its target transcripts. The convincing results demonstrate, with multiple independent approaches, co-transcriptional binding, stabilization of a family of mRNAs, and cytoplasmic activities of the transcription factor Sfp1. The results lead to the conclusion that the co-transcriptional association of Sfp1 with specific transcripts is a critical step in the stabilization of such transcripts in the cytoplasm.
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
To function effectively as an integrated system, the transcriptional and post-transcriptional machineries must communicate through mechanisms that are still poorly understood. Here, we focus on the zinc-finger Sfp1, known to regulate transcription of proliferation-related genes. We show that Sfp1 can regulate transcription either by binding to promoters, like most known transcription activators, or by binding to the transcribed regions (gene bodies), probably via RNA polymerase II (Pol II). We further studied the first mode of Sfp1 activity and found that, following promoter binding, Sfp1 binds to gene bodies and affects Pol II configuration, manifested by dissociation or conformational change of its Rpb4 subunit and increased backtracking. Surprisingly, Sfp1 binds to a subset of mRNAs co-transcriptionally and stabilizes them. The interaction between Sfp1 and its client mRNAs is controlled by their respective promoters and coincides with Sfp1’s dissociation from chromatin. Intriguingly, Sfp1 dissociation from the chromatin correlates with the extent of the backtracked Pol II. We propose that, following promoter recruitment, Sfp1 accompanies Pol II and regulates backtracking. The backtracked Pol II is more compatible with Sfp1’s relocation to the nascent transcripts, whereupon Sfp1 accompanies these mRNAs to the cytoplasm and regulates their stability. Thus, Sfp1’s co-transcriptional binding imprints the mRNA fate, serving as a paradigm for the cross-talk between the synthesis and decay of specific mRNAs, and a paradigm for the dual-role of some zinc-finger proteins. The interplay between Sfp1’s two modes of transcription regulation remains to be examined.
Article activity feed
-

-

-

-

eLife assessment
This important study reports that a transcription factor stimulating mRNA synthesis can stabilize its target transcripts. The convincing results demonstrate, with multiple independent approaches, co-transcriptional binding, stabilization of a family of mRNAs, and cytoplasmic activities of the transcription factor Sfp1. The results lead to the conclusion that the co-transcriptional association of Sfp1 with specific transcripts is a critical step in the stabilization of such transcripts in the cytoplasm.
-

Reviewer #1 (Public Review):
This manuscript builds upon the authors' previous work on the cross-talk between transcription initiation and post-transcriptional events in yeast gene expression. These prior studies identified an mRNA 'imprinting' phenomenon linked to genes activated by the Rap1 transcription factor (TF), a surprising role for the Sfp1 TF in promoting RNA polymerase II (RNAPII) backtracking, and a role for the non-essential RNAPII subunits Rpb4/7 in the regulation of mRNA decay and translation. Here the authors aimed to extend these observations to provide a more coherent picture of the role of Sfp1 in transcription initiation and subsequent steps in gene expression. They provide evidence for (1) a physical interaction between Sfp1 and Rpb4, (2) Sfp1 binding and stabilization of mRNAs derived from genes whose promoters are …
Reviewer #1 (Public Review):
This manuscript builds upon the authors' previous work on the cross-talk between transcription initiation and post-transcriptional events in yeast gene expression. These prior studies identified an mRNA 'imprinting' phenomenon linked to genes activated by the Rap1 transcription factor (TF), a surprising role for the Sfp1 TF in promoting RNA polymerase II (RNAPII) backtracking, and a role for the non-essential RNAPII subunits Rpb4/7 in the regulation of mRNA decay and translation. Here the authors aimed to extend these observations to provide a more coherent picture of the role of Sfp1 in transcription initiation and subsequent steps in gene expression. They provide evidence for (1) a physical interaction between Sfp1 and Rpb4, (2) Sfp1 binding and stabilization of mRNAs derived from genes whose promoters are bound by both Rap1 and Sfp1 and (3) an effect of Sfp1 on Rpb4 binding or conformation during transcription elongation.
-

Author response:
The following is the authors’ response to the previous reviews.
Public Reviews:
Reviewer #2 (Public Review):
Summary:
The manuscript by Kelbert et al. presents results on the involvement of the yeast transcription factor Sfp1 in the stabilisation of transcripts whose synthesis it stimulates. Sfp1 is known to affect the synthesis of a number of important cellular transcripts, such as many of those that code for ribosomal proteins. The hypothesis that a transcription factor can remain bound to the nascent transcript and affect its cytoplasmic half-life is attractive. However, the association of Sfp1 with cytoplasmic transcripts remains to be validated, as explained in the following comments:
A two-hybrid based assay for protein-protein interactions identified Sfp1, a transcription factor known for its effects on …
Author response:
The following is the authors’ response to the previous reviews.
Public Reviews:
Reviewer #2 (Public Review):
Summary:
The manuscript by Kelbert et al. presents results on the involvement of the yeast transcription factor Sfp1 in the stabilisation of transcripts whose synthesis it stimulates. Sfp1 is known to affect the synthesis of a number of important cellular transcripts, such as many of those that code for ribosomal proteins. The hypothesis that a transcription factor can remain bound to the nascent transcript and affect its cytoplasmic half-life is attractive. However, the association of Sfp1 with cytoplasmic transcripts remains to be validated, as explained in the following comments:
A two-hybrid based assay for protein-protein interactions identified Sfp1, a transcription factor known for its effects on ribosomal protein gene expression, as interacting with Rpb4, a subunit of RNA polymerase II. Classical two-hybrid experiments depend on the presence of the tested proteins in the nucleus of yeast cells, suggesting that the observed interaction occurs in the nucleus. Unfortunately, the two-hybrid method cannot determine whether the interaction is direct or mediated by nucleic acids. The revised version of the manuscript now states that the observed interaction could be indirect.
To understand to which RNA Sfp1 might bind, the authors used an N-terminally tagged fusion protein in a cross-linking and purification experiment. This method identified 264 transcripts for which the CRAC signal was considered positive and which mostly correspond to abundant mRNAs, including 74 ribosomal protein mRNAs or metabolic enzyme-abundant mRNAs such as PGK1. The authors did not provide evidence for the specificity of the observed CRAC signal, in particular what would be the background of a similar experiment performed without UV cross-linking. This is crucial, as Figure S2G shows very localized and sharp peaks for the CRAC signal, often associated with over-amplification of weak signal during sequencing library preparation.
(1) To rule out possible PCR artifacts, we used a UMI (Unique Molecular Identifier) scan. UMIs are short, random sequences added to each molecule by the 5’ adapter to uniquely tag them. After PCR amplification and alignment to the reference genome, groups of reads with identical UMIs represent only one unique original molecule. Thus, UMIs allow distinguishing between original molecules and PCR duplicates, effectively eliminating the duplicates.
(2) Looking closely at the peaks using the IGV browser, we noticed that the reads are by no means identical. Each carrying a mutation [probably due to the cross-linking] in a different position and having different length. Note that the reads are highly reproducible in two replicate.
(3) CRAC+ genes do not all fall into the category of highly transcribed genes. On the contrary, as depicted in Figure 6A (green dots), it is evident that CRAC+ genes exhibit a diverse range of Rpb3 ChIP and GRO signals. Furthermore, as illustrated in Figure 7A, when comparing CRAC+ to Q1 (the most highly transcribed genes), it becomes evident that the Rpb4/Rpb3 profile of CRAC+ genes is not a result of high transcription levels.
(4) Only a portion of the RiBi mRNAs binds Sfp1, despite similar expression of all RiBi.
(5) The CRAC+ genes represent a distinct group with many unique features. Moreover, many CRAC+ genes do not fall into the category of highly transcribed genes.
(6) The biological significance of the 262 CRAC+ mRNAs was demonstrated by various experiments; all are inconsistent with technical flaws. Some examples are:
a) Fig. 2a and B show that most reads of CRAC+ mRNA were mapped to specific location – close the pA sites.
b) Fig. 2C shows that most reads of CRAC+ mRNA were mapped to specific RNA motif.
c) Most RiBi CRAC+ promoter contain Rap1 binding sites (p= 1.9x10-22), whereas the vast majority of RiBi CRAC- promoters do not contain Rap1 binding site. (Fig. 3C).
d) Fig. 4A shows that RiBi CRAC+ mRNAs become destabilized due to Sfp1 deletion, whereas RiBi CRAC- mRNAs do not. Fig. 4B shows similar results due to
e) Fig. 6B shows that the impact of Sfp1 on backtracking is substantially higher for CRAC+ than for CRAC- genes. This is most clearly visible in RiBi genes.
f) Fig. 7A shows that the Sfp1-dependent changes along the transcription units is substantially more rigorous for CRAC+ than for CRAC-.
g) Fig. S4B Shows that chromatin binding profile of Sfp1 is different for CRAC+ and CRAC- genes
In a validation experiment, the presence of several mRNAs in a purified SFP1 fraction was measured at levels that reflect the relative levels of RNA in a total RNA extract. Negative controls showing that abundant mRNAs not found in the CRAC experiment were clearly depleted from the purified fraction with Sfp1 would be crucial to assess the specificity of the observed protein-RNA interactions (to complement Fig. 2D).
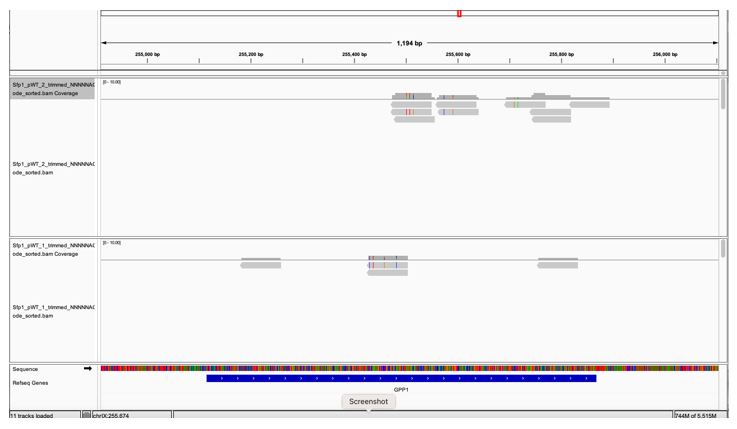
GPP1, a highly expressed genes, is not to be pulled down by Sfp1 (Fig. 2D). GPP1 (alias RHR2) was included in our Table S2 as one of the 264 CRAC+ genes, having a low CRAC value. However, when we inspected GPP1 results using the IGV browser, we realized that the few reads mapped to GPP1 are actually anti-sense to GPP1 (perhaps they belong to the neighboring RPL34B genes, which is convergently transcribed to GPP1) (see Fig. 1 at the bottom of the document). Thus, GPP1 is not a CRAC+ gene and would now serve as a control. See We changed the text accordingly (see page 11 blue sentences). In light of this observation, we checked other CRAC genes and found that, except for ALG2, they all contain sense reads (some contain both sense and anti-sense reads). ALG2 and GPP1 were removed leaving 262 CRAC+ genes.
The CRAC-selected mRNAs were enriched for genes whose expression was previously shown to be upregulated upon Sfp1 overexpression (Albert et al., 2019). The presence of unspliced RPL30 pre-mRNA in the Sfp1 purification was interpreted as a sign of co-transcriptional assembly of Sfp1 into mRNA, but in the absence of valid negative controls, this hypothesis would require further experimental validation. Also, whether the fraction of mRNA bound by Sfp1 is nuclear or cytoplasmic is unclear.
Further experimental validation was provided in some of our figures (e.g., Fig. 5C, Fig. 3B).
We argue that Sfp1 binds RNA co-transcriptionally and accompanies the mRNA till its demise in the cytoplasm: Co-transcriptional binding is shown in: (I) a drop in the Sfp1 ChIP-exo signal that coincides with the position of Sfp1 binding site in the RNA (Fig. 5C), demonstrating a movement of Sfp1 from chromatin to the transcript, (II) the dependence of Sfp1 RNA-binding on the promoter (Fig. 3B) and binding of intron-containing RNA. Taken together these 3 different experiments demonstrate that Sfp1 binds Pol II transcript co-transcriptionally. Association of Sfp1 with cytoplasmic mRNAs is shown in the following experiments: (I) Figure 2D shows that Sfp1 pulled down full length RNA, strongly suggesting that these RNA are mature cytoplasmic mRNAs. (II) mRNA encoding ribosomal proteins, which belong to the CRAC+ mRNAs group are degraded by Xrn1 in the cytoplasm (Bresson et al., Mol Cell 2020). The capacity of Sfp1 to regulates this process (Fig. 4A-D) is therefore consistent with cytoplasmic activity of Sfp1. (III) The effect of Sfp1 on deadenylation (Fig. 4D), a cytoplasmic process, is also consistent with cytoplasmic activity of Sfp1.
To address the important question of whether co-transcriptional assembly of Spf1 with transcripts could alter their stability, the authors first used a reporter system in which the RPL30 transcription unit is transferred to vectors under different transcriptional contexts, as previously described by the Choder laboratory (Bregman et al. 2011). While RPL30 expressed under an ACT1 promoter was barely detectable, the highest levels of RNA were observed in the context of the native upstream RPL30 sequence when Rap1 binding sites were also present. Sfp1 showed better association with reporter mRNAs containing Rap1 binding sites in the promoter region. Removal of the Rap1 binding sites from the reporter vector also led to a drastic decrease in reporter mRNA levels. Co-purification of reporter RNA with Sfp1 was only observed when Rap1 binding sites were included in the reporter. Negative controls for all the purification experiments might be useful.
In the swapping experiment, the plasmid lacking RapBS serves as the control for the one with RapBS and vice versa (see Bregman et al., 2011). Remember, that all these contracts give rise to identical RNA. Indeed, RabBS affects both mRNA synthesis and decay, therefore the controls are not ideal. However, see next section.
More importantly, in Fig. 3B “Input” panel, one can see that the RNA level of “construct F” was higher than the level of “construct E”. Despite this difference, only the RNA encoded by construct E was detected in the IP panel. This clearly shows that the detection of the RNA was not merely a result of its expression level.
To complement the biochemical data presented in the first part of the manuscript, the authors turned to the deletion or rapid depletion of SFP1 and used labelling experiments to assess changes in the rate of synthesis, abundance and decay of mRNAs under these conditions. An important observation was that in the absence of Sfp1, mRNAs encoding ribosomal protein genes not only had a reduced synthesis rate, but also an increased degradation rate. This important observation needs careful validation,
Indeed, we do provide validations in Fig. 4C Fig. 4D Fig. S3A and during the revision we included an additional validation as Fig. S3B. Of note, we strongly suspect that GRO is among the most reliable approaches to determine half-lives (see our response in the first revision letter).
As genomic run-on experiments were used to measure half-lives, and this particular method was found to give results that correlated poorly with other measures of half-life in yeast (e.g. Chappelboim et al., 2022 for a comparison). As an additional validation, a temperature shift to 42{degree sign}C was used to show that , for specific ribosomal protein mRNA, the degradation was faster, assuming that transcription stops at that temperature. It would be important to cite and discuss the work from the Tollervey laboratory showing that a temperature shift to 42{degree sign}C leads to a strong and specific decrease in ribosomal protein mRNA levels, probably through an accelerated RNA degradation (Bresson et al., Mol Cell 2020, e.g. Fig 5E).
This was cited. Thank you.
Finally, the conclusion that mRNA deadenylation rate is altered in the absence of Sfp1, is difficult to assess from the presented results (Fig. 3D).
This type of experiment was popular in the past. The results in the literature are similar to ours (in fact, ours are nicer). Please check the papers cited in our MS and a number of papers by Roy Parker.
The effects of SFP1 on transcription were investigated by chromatin purification with Rpb3, a subunit of RNA polymerase, and the results were compared with synthesis rates determined by genomic run-on experiments. The decrease in polII presence on transcripts in the absence of SFP1 was not accompanied by a marked decrease in transcript output, suggesting an effect of Sfp1 in ensuring robust transcription and avoiding RNA polymerase backtracking. To further investigate the phenotypes associated with the depletion or absence of Sfp1, the authors examined the presence of Rpb4 along transcription units compared to Rpb3. An effect of spf1 deficiency was that this ratio, which decreased from the start of transcription towards the end of transcripts, increased slightly. To what extent this result is important for the main message of the manuscript is unclear.
Suggestions: a) please clearly indicate in the figures when they correspond to reanalyses of published results.
This was done.
b) In table S2, it would be important to mention what the results represent and what statistics were used for the selection of "positive" hits.
This was discussed in the text.
Strengths:
- Diversity of experimental approaches used.
- Validation of large-scale results with appropriate reporters.
Weaknesses:
- Lack of controls for the CRAC results and lack of negative controls for the co-purification experiments that were used to validate specific mRNA targets potentially bound by Sfp1.
- Several conclusions are derived from complex correlative analyses that fully depend on the validity of the aforementioned Sfp1-mRNA interactions.
We hope that our responses to Reviewer 2's thoughtful comments have rulled out concerns regarding the lack of controls.
Recommendations for the authors:
Reviewer #2 (Recommendations For The Authors):
Please review the text for spelling errors. While not mandatory, wig or begraph files for the CRAC results would be very useful for the readers.
Author response image 1.
A snapshot of IGV GPP1 locus showing that all the reads are anti-sense (pointing at the opposite direction of the gene (the gene arrows [white arrows over blue, at the bottom] are pointing to the right whereas the reads’ orientations are pointing to the left).
-

-

eLife assessment
This important study reports that a transcription factor that stimulates mRNA synthesis can stabilize its target transcripts, possibly through co-transcriptional assembly and action in the cytoplasm. While the primary observation is solid, whether an association of Sfp1 with specific transcripts in the cytoplasm is the critical step in transcript stabilization is not entirely clear. If confirmed by independent means, the authors would have found a novel mechanistic link between mRNA synthesis and cytoplasmic mRNA stability for specific transcripts. Such a finding would be of broad interest to the field of molecular biology.
-

Reviewer #2 (Public Review):
Summary:
The manuscript by Kelbert et al. presents results on the involvement of the yeast transcription factor Sfp1 in the stabilisation of transcripts whose synthesis it stimulates. Sfp1 is known to affect the synthesis of a number of important cellular transcripts, such as many of those that code for ribosomal proteins. The hypothesis that a transcription factor can remain bound to the nascent transcript and affect its cytoplasmic half-life is attractive. However, the association of Sfp1 with cytoplasmic transcripts remains to be validated, as explained in the following comments:
A two-hybrid based assay for protein-protein interactions identified Sfp1, a transcription factor known for its effects on ribosomal protein gene expression, as interacting with Rpb4, a subunit of RNA polymerase II. Classical …
Reviewer #2 (Public Review):
Summary:
The manuscript by Kelbert et al. presents results on the involvement of the yeast transcription factor Sfp1 in the stabilisation of transcripts whose synthesis it stimulates. Sfp1 is known to affect the synthesis of a number of important cellular transcripts, such as many of those that code for ribosomal proteins. The hypothesis that a transcription factor can remain bound to the nascent transcript and affect its cytoplasmic half-life is attractive. However, the association of Sfp1 with cytoplasmic transcripts remains to be validated, as explained in the following comments:
A two-hybrid based assay for protein-protein interactions identified Sfp1, a transcription factor known for its effects on ribosomal protein gene expression, as interacting with Rpb4, a subunit of RNA polymerase II. Classical two-hybrid experiments depend on the presence of the tested proteins in the nucleus of yeast cells, suggesting that the observed interaction occurs in the nucleus. Unfortunately, the two-hybrid method cannot determine whether the interaction is direct or mediated by nucleic acids. The revised version of the manuscript now states that the observed interaction could be indirect.
To understand to which RNA Sfp1 might bind, the authors used an N-terminally tagged fusion protein in a cross-linking and purification experiment. This method identified 264 transcripts for which the CRAC signal was considered positive and which mostly correspond to abundant mRNAs, including 74 ribosomal protein mRNAs or metabolic enzyme-abundant mRNAs such as PGK1. The authors did not provide evidence for the specificity of the observed CRAC signal, in particular what would be the background of a similar experiment performed without UV cross-linking. This is crucial, as Figure S2G shows very localized and sharp peaks for the CRAC signal, often associated with over-amplification of weak signal during sequencing library preparation.
In a validation experiment, the presence of several mRNAs in a purified SFP1 fraction was measured at levels that reflect the relative levels of RNA in a total RNA extract. Negative controls showing that abundant mRNAs not found in the CRAC experiment were clearly depleted from the purified fraction with Sfp1 would be crucial to assess the specificity of the observed protein-RNA interactions (to complement Fig. 2D). The CRAC-selected mRNAs were enriched for genes whose expression was previously shown to be upregulated upon Sfp1 overexpression (Albert et al., 2019). The presence of unspliced RPL30 pre-mRNA in the Sfp1 purification was interpreted as a sign of co-transcriptional assembly of Sfp1 into mRNA, but in the absence of valid negative controls, this hypothesis would require further experimental validation. Also, whether the fraction of mRNA bound by Sfp1 is nuclear or cytoplasmic is unclear.
To address the important question of whether co-transcriptional assembly of Spf1 with transcripts could alter their stability, the authors first used a reporter system in which the RPL30 transcription unit is transferred to vectors under different transcriptional contexts, as previously described by the Choder laboratory (Bregman et al. 2011). While RPL30 expressed under an ACT1 promoter was barely detectable, the highest levels of RNA were observed in the context of the native upstream RPL30 sequence when Rap1 binding sites were also present. Sfp1 showed better association with reporter mRNAs containing Rap1 binding sites in the promoter region. Removal of the Rap1 binding sites from the reporter vector also led to a drastic decrease in reporter mRNA levels. Co-purification of reporter RNA with Sfp1 was only observed when Rap1 binding sites were included in the reporter. Negative controls for all the purification experiments might be useful.
To complement the biochemical data presented in the first part of the manuscript, the authors turned to the deletion or rapid depletion of SFP1 and used labelling experiments to assess changes in the rate of synthesis, abundance and decay of mRNAs under these conditions. An important observation was that in the absence of Sfp1, mRNAs encoding ribosomal protein genes not only had a reduced synthesis rate, but also an increased degradation rate. This important observation needs careful validation, as genomic run-on experiments were used to measure half-lives, and this particular method was found to give results that correlated poorly with other measures of half-life in yeast (e.g. Chappelboim et al., 2022 for a comparison). As an additional validation, a temperature shift to 42{degree sign}C was used to show that , for specific ribosomal protein mRNA, the degradation was faster, assuming that transcription stops at that temperature. It would be important to cite and discuss the work from the Tollervey laboratory showing that a temperature shift to 42{degree sign}C leads to a strong and specific decrease in ribosomal protein mRNA levels, probably through an accelerated RNA degradation (Bresson et al., Mol Cell 2020, e.g. Fig 5E). Finally, the conclusion that mRNA deadenylation rate is altered in the absence of Sfp1, is difficult to assess from the presented results (Fig. 3D).
The effects of SFP1 on transcription were investigated by chromatin purification with Rpb3, a subunit of RNA polymerase, and the results were compared with synthesis rates determined by genomic run-on experiments. The decrease in polII presence on transcripts in the absence of SFP1 was not accompanied by a marked decrease in transcript output, suggesting an effect of Sfp1 in ensuring robust transcription and avoiding RNA polymerase backtracking. To further investigate the phenotypes associated with the depletion or absence of Sfp1, the authors examined the presence of Rpb4 along transcription units compared to Rpb3. An effect of spf1 deficiency was that this ratio, which decreased from the start of transcription towards the end of transcripts, increased slightly. To what extent this result is important for the main message of the manuscript is unclear.
Suggestions: a) please clearly indicate in the figures when they correspond to reanalyses of published results. b) In table S2, it would be important to mention what the results represent and what statistics were used for the selection of "positive" hits.
Strengths:
- Diversity of experimental approaches used.
- Validation of large-scale results with appropriate reporters.Weaknesses:
- Lack of controls for the CRAC results and lack of negative controls for the co-purification experiments that were used to validate specific mRNA targets potentially bound by Sfp1.
- Several conclusions are derived from complex correlative analyses that fully depend on the validity of the aforementioned Sfp1-mRNA interactions. -

Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public Review):
Summary:
This manuscript builds upon the authors' previous work on the cross-talk between transcription initiation and post-transcriptional events in yeast gene expression. These prior studies identified an mRNA 'imprinting' phenomenon linked to genes activated by the Rap1 transcription factor (TF), a surprising role for the Sfp1 TF in promoting RNA polymerase II (RNAPII) backtracking, and a role for the non-essential RNAPII subunits Rpb4/7 in the regulation of mRNA decay and translation. Here the authors aimed to extend these observations to provide a more coherent picture of the role of Sfp1 in transcription initiation and subsequent steps in gene expression. They provide evidence for (1) a physical interaction between Sfp1 …
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public Review):
Summary:
This manuscript builds upon the authors' previous work on the cross-talk between transcription initiation and post-transcriptional events in yeast gene expression. These prior studies identified an mRNA 'imprinting' phenomenon linked to genes activated by the Rap1 transcription factor (TF), a surprising role for the Sfp1 TF in promoting RNA polymerase II (RNAPII) backtracking, and a role for the non-essential RNAPII subunits Rpb4/7 in the regulation of mRNA decay and translation. Here the authors aimed to extend these observations to provide a more coherent picture of the role of Sfp1 in transcription initiation and subsequent steps in gene expression. They provide evidence for (1) a physical interaction between Sfp1 and Rpb4, (2) Sfp1 binding and stabilization of mRNAs derived from genes whose promoters are bound by both Rap1 and Sfp1 and (3) an effect of Sfp1 on Rpb4 binding or conformation during transcription elongation.
Strengths:
This study provides evidence that a TF (yeast Sfp1), in addition to stimulating transcription initiation, can at some target genes interact with their mRNA transcripts and promote their stability. Sfp1 thus has a positive effect on two distinct regulatory steps. Furthermore, evidence is presented indicating that strong Sfp1 mRNA association requires both Rap1 and Sfp1 promoter binding and is increased at a sequence motif near the polyA track of many target mRNAs. Finally, they provide compelling evidence that Sfp1-bound mRNAs have higher levels of RNAPII backtracking and altered Rpb4 association or conformation compared to those not bound by Sfp1.
Weaknesses:
The Sfp1-Rpb4 association is supported only by a two-hybrid assay that is poorly described and lacks an important control. Furthermore, there is no evidence that this interaction is direct, nor are the interaction domains on either protein identified (or mutated to address function).
Indeed, our two hybrid, immunoprecipitation and imaging results do not allow us to conclusively discern whether the interaction between Rpb4 and Sfp1 is direct or indirect. While the interaction holds significance, we consider the direct versus indirect distinction to be of secondary importance in the context of this paper. In the current text we indicated that 'our two hybrid, immunoprecipitation and imaging results do not differentiate between a direct or indirect interactions' (see page 6, sentences highlighted in blue)
The contention that Sfp1 nuclear export to the cytoplasm is transcription-dependent is not well supported by the experiments shown, which are not properly described in the text and are not accompanied by any primary data.
This section has been re-written for better clarity (see page 7). We note that this assay was originally developed and published by Lee, M. S., M. Henry, and P. A. Silver in their 1996 paper in G&D and has since been reported in numerous subsequent studies. Reassuringly, our conclusion is bolstered by the observation that Sfp1 binds to Pol II transcripts co-transcriptionally, suggesting that Sfp1 is exported in the context of the mRNA.
The presence of Sfp1 in P-bodies is of unclear relevance and the authors do not ask whether Sfp1-bound mRNAs are also present in these condensates.
P-bodies consist of both RNA and proteins (reviewed in doi: 10.1021/acs.biochem.7b01162). The significance of this experiment lies in its contribution to further confirming the co-localization of Sfp1 with mRNAs and Rpb4. This observation could also yield valuable insights for future investigations into the role of Sfp1.
Further analysis of Sfp1-bound mRNAs would be of interest, particularly to address the question of whether those from ribosomal protein genes and other growth-related genes that are known to display Sfp1 binding in their promoters are regulated (either stabilized or destabilized) by Sfp1.
Fig. 4A, C and D show that RP mRNAs become destabilized in sfp1Δ cells.
The authors need to discuss, and ideally address, the apparent paradox that their previous findings showed that Rap1 acts to destabilize its downstream transcripts, i.e. that it has the opposite effect of Sfp1 shown here.
We would like to thank Reviewer 1 for this valuable comment. In the revised paper, we delved into our hypothesis suggesting that Rap1 is likely responsible for regulating the imprinting of other proteins, that, in turn, lead to the destabilization of mRNAs, such as Rpb4. See blue paragraph in page 20.
Finally, recent studies indicate that the drugs used here to measure mRNA stability induce a strong stress response accompanied by rapid and complex effects on transcription. Their relevance to mRNA stability in unstressed cells is questionable.
Half-lives were determined mainly by the GRO analysis of optimally proliferating cells. This method does not requires any drug or stressful treatment. The results obtained by this method were consistent with those obtained after thiolutin addition. Using both methods, we discovered that disruption of Sfp1 results in substantial mRNA destabilization. Nevertheless, in our revised manuscript, we show results obtained by subjecting cells to a temperature shift to 42°C, a natural method to inhibit transcription. This approach to determine half-lives has been previously reported in our publications, such as Lotan et al. (2005, 2007) and Goler Baron et al. (2008). This may rule out effects of the drug on half-lives. Indeed, this assay clearly determine HL under heat stress. Thus it can clearly demonstrate that, at least during heat shock, Sfp1 stabilizes mRNAs. Since the results are similar to those obtained by the GRO method at 30oC, we concluded that Sfp1 stabilizes mRNA under optimal and hot conditions.
Reviewer #2 (Public Review):
Summary:
The manuscript by Kelbert et al. presents results on the involvement of the yeast transcription factor Sfp1 in the stabilisation of transcripts whose synthesis it stimulates. Sfp1 is known to affect the synthesis of a number of important cellular transcripts, such as many of those that code for ribosomal proteins. The hypothesis that a transcription factor can remain bound to the nascent transcript and affect its cytoplasmic half-life is attractive, but the methods used to demonstrate the half-life effects and the association of Sfp1 with cytoplasmic transcripts remain to be fully validated, as explained in my comments on the results below:
Comments on methodology and results:
(1) A two-hybrid-based assay for protein-protein interactions identified Sfp1, a transcription factor known for its effects on ribosomal protein gene expression, as interacting with Rpb4, a subunit of RNA polymerase II. Classical two-hybrid experiments depend on the presence of the tested proteins in the nucleus of yeast cells, suggesting that the observed interaction occurs in the nucleus. Unfortunately, the two-hybrid method cannot determine whether the interaction is direct or mediated by nucleic acids.
Indeed, our two hybrid, immunoprecipitation and imaging results do not allow us to conclusively discern whether the interaction between Rpb4 and Sfp1 is direct or indirect. While the interaction holds significance, we consider the direct versus indirect distinction to be of secondary importance in the context of this paper. In the current text we indicated that 'our two hybrid, immunoprecipitation and imaging results do not differentiate between a direct or indirect interactions' (see page 6)
(2) Inactivation of nup49, a component of the nuclear pore complex, resulted in the redistribution of GFP-Sfp1 into the cytoplasm at the temperature non-permissive for the nup49-313 strain, suggesting that GFP-Sfp1 is a nucleo-cytoplasmic shuttling protein. This observation confirmed the dynamic nature of the nucleo-cytoplasmic distribution of Sfp1. For example, a similar redistribution to the cytoplasm was previously reported following rapamycin treatment and under starvation (Marion et al., PNAS 2004). In conjunction with the observation of an interaction with Rpb4, the authors observed slower nuclear import kinetics for GFP-Sfp1 in the absence of Rpb4 when cells were transferred to a glucose-containing medium after a period of starvation. Since the redistribution of GFP-Sfp1 was abolished in an rpb1-1/nup49-313 double mutant, the authors concluded that Sfp1 localisation to the cytoplasm depends on transcription. The double mutant yeast cells may show a variety of non-specific effects at the restrictive temperature, and whether transcription is required for Sfp1 cytoplasmic localisation remains incompletely demonstrated.
We agree with Reviewer 2 that any heat inactivation of a temperature-sensitive (ts) protein can lead to non-specific effects. It is evident that nup49-313 does not prevent Sfp1 export to the cytoplasm. In the case of rpb1-1, these non-specific effects are expected due to transcriptional arrest, which can eventually result in a reduction in protein content. However, this process takes some time, while the impact on export is more rapid. It is worth noting that this assay was developed and previously published by Pam Silver (Henry and Silver G&D 1996) and has been reported in many subsequent papers. Importantly, our conclusion is supported by the observation that Sfp1 binds both nascent RNA (co-transcriptionally) and mature mRNA (cytoplasmic). These observations, along with the reduced mRNA export upon transcription blocking, are consistent with our proposal that Sfp1 is exported in association with mRNA.
(3) Under starvation conditions, which led to the presence of Sfp1 in the cytoplasm and have previously been correlated with a decrease in the transcription of Sfp1 target genes, the authors observed that a plasmid-based expressed GFP-Sfp1 accumulated in cytoplasmic foci. These foci were also labelled by P-body markers such as Dcp2 and Lsm1. The quality of the microscopic images provided does not allow to determine whether Rpb4-RFP colocalises with GFP-Sfp1.
The submitted PDF figure is of low quality. We believe that high quality figure of the final submission is convincing.
(4) To understand to which RNA Sfp1 might bind, the authors used an N-terminally tagged fusion protein in a cross-linking and purification experiment. This method identified 264 transcripts for which the CRAC signal was considered positive and which mostly correspond to abundant mRNAs, including 74 ribosomal protein mRNAs or metabolic enzyme-abundant mRNAs such as PGK1. The authors did not provide evidence for the specificity of the observed CRAC signal, in particular, what would be the background of a similar experiment performed without UV cross-linking. In a validation experiment, the presence of several mRNAs in a purified SFP1 fraction was measured at levels that reflect the relative levels of RNA in a total RNA extract. Negative controls showing that abundant mRNAs not found in the CRAC experiment were clearly depleted from the purified fraction with Sfp1 would be crucial to assessing the specificity of the observed protein-RNA interactions. The NON-CRAC+ selected mRNAs were enriched for genes whose expression was previously shown to be upregulated upon Sfp1 overexpression (Albert et al., 2019). The presence of unspliced RPL30 pre-mRNA in the Sfp1 purification was interpreted as a sign of co-transcriptional assembly of Sfp1 into mRNA, but in the absence of valid negative controls, this hypothesis would require further experimental validation.
We would like to thank Reviewer 2 for bringing this issue up, as it helped us to clarify it in the revised paper.
First, we emphasized in the Discussion that many CRAC+ genes do not fall into the category of highly transcribed genes. Please see more detailed discussion below.
Secondly, we examined various features of the 264 genes - classified as CRAC+ - to estimate their specificity and biological significance. Our various experiments revealed that the CRAC+ genes represent a distinct group with many unique features.
The biological significance of the 264 CRAC+ mRNAs was demonstrated by various experiments; all are inconsistent with technical flaws. In fact, all the experiments and analyses that we have pursued indicate the unique nature of the CRAC+ genes. Some examples are:
(1) Fig. 2a and B show that most reads of CRAC+ mRNA were mapped to specific location – close the pA sites.
(2) Fig. 2C shows that most reads of CRAC+ mRNA were mapped to specific RNA motif located near the 3’ ends of the mRNAs.
(3) Most RiBi CRAC+ promoter contain Rap1 binding sites (p= 1.9x10-22), whiles the vast majority of RiBi non-CRAC+ promoters do not. (Fig. 3C).
(4) Fig. 4A shows that RiBi CRAC+ mRNAs become destabilized due to Sfp1 deletion, whereas RiBi non-CRAC+ mRNAs do not. Fig. 4B shows similar results due to Sfp1 depletion.
(5) Fig. 6B shows that the impact of Sfp1 on backtracking is substantially higher for CRAC+ than for non-CRAC+ genes. This is most clearly visible in RiBi genes.
(6) Fig. 7A shows that the Sfp1-dependent changes along the transcription units is substantially more rigorous for CRAC+ than for non-CRAC+.
(7) In Fig. S4B, the chromatin binding profile of Sfp1 is shown to be different for CRAC+ and non-CRAC+ genes.
Taken together, the many unique features, in fact, any feature that we examined, indicate the specificity and significance of this group, demonstrating that our CRAC results are biologically significant.
Most importantly, these genes do not all fall into the category of highly transcribed genes. On the contrary, as depicted in Figure 6A (green dots), it is evident that CRAC+ genes exhibit a diverse range of Rpb3 ChIP and GRO signals. Furthermore, as illustrated in Figure 7A, when comparing CRAC+ to Q1 (the most highly transcribed genes), it becomes evident that the Rpb4/Rpb3 profile of CRAC+ genes behaves differently from the Q1 group. Evidently, despite the heterogeneous transcription of CRAC+ genes (as mentioned above), the Rpb4/Rpb3 profile decreases more substantially than that of the highly transcribed genes (Q1). Moreover, despite similar expression levels among all RiBi mRNAs, only a portion of them binds Sfp1.
Thus, all our results indicate that CRAC+ genes represent biologically significant group, irrespective of the expression of it members. In response to this comment, we included a new paragraph discussing the validity of our conclusions. See page 18, blue paragraph.
(5) To address the important question of whether co-transcriptional assembly of Spf1 with transcripts could alter their stability, the authors first used a reporter system in which the RPL30 transcription unit is transferred to vectors under different transcriptional contexts, as previously described by the Choder laboratory (Bregman et al. 2011). While RPL30 expressed under an ACT1 promoter was barely detectable, the highest levels of RNA were observed in the context of the native upstream RPL30 sequence when Rap1 binding sites were also present. Sfp1 showed better association with reporter mRNAs containing Rap1 binding sites in the promoter region. However, removal of the Rap1 binding sites from the reporter vector also led to a drastic decrease in reporter mRNA levels. Whether the fraction of co-purified RNA is nuclear and co-transcriptional or not cannot be inferred from these results.
The proposed co-transcriptional binding of Sfp1 is based on the findings presented in Figure 5C and Figure S2D, as well as the observed binding of Sfp1 to transcripts containing introns, as shown in Figures 2D and 3B. The results of Fig. 3 led us to the assertion that the "RNA-binding capacity of Sfp1 is regulated by Rap1-binding sites located at the promoter." We maintain our stance on this conclusion. Indeed, the Rap1 binding site does impact mRNA levels, as highlighted by Reviewer 2. However, "construct E," which possesses a promoter with a Rap1 binding site, exhibits lower transcript levels compared to "construct F," which lacks such a binding site in its promoter. Despite this difference in transcript levels, Sfp1 was able to pull down the former transcript but not the latter, even though expression of the former gene is relatively low. Thus, the results appear to be more reliant on the specific capacity of Sfp1 to interact with the transcript rather than on the transcript's expression level.
(6) To complement the biochemical data presented in the first part of the manuscript, the authors turned to the deletion or rapid depletion of SFP1 and used labelling experiments to assess changes in the rate of synthesis, abundance, and decay of mRNAs under these conditions. An important observation was that in the absence of Sfp1, mRNAs encoding ribosomal protein genes not only had a reduced synthesis rate but also an increased degradation rate. This important observation needs careful validation, as genomic run-on experiments were used to measure half-lives, and this particular method was found to give results that correlated poorly with other measures of half-life in yeast (e.g. Chappelboim et al., 2022 for a comparison). Similarly, the use of thiolutin to block transcription as a method of assessing mRNA half-life has been reported to be problematic, as thiolutin can specifically inhibit the degradation of ribosomal protein mRNA (Pelechano & Perez-Ortin, 2008). Specific repressible reporters, such as those used by Baudrimont et al. (2017), would need to be tested to validate the effect of Sfp1 on the half-life of specific mRNAs. Also, it would be very difficult to infer from the images presented whether the rate of deadenylation is altered by Sfp1.
Various methods exist for assessing mRNA half-lives (HLs), and each of them carries its own set of challenges and biases. Consequently, it becomes problematic to directly compare HL values of a specific mRNA when different methods are employed. The superiority of one particular method over others remains unclear (in my opinion). However, they exhibit a high degree of reliability when it comes to comparing different strains under the identical conditions using a single method.
Estimating HLs through the GRO approach is a non-invasive method, applied on optimally proliferating cells, which has been employed in numerous publications. While no method is without its limitations, our experience along the years reassured approach to be among the most dependable. Our HL determination using thiolutin to block transcription provided results that were consistent with the values obtained by the GRO approach.
Nevertheless, in our revised manuscript, we supplemented the HL data, obtain by thiolutin, with results obtained by subjecting cells to a temperature shift to 42°C, a natural method to block transcription in wild-type (WT) cells. This approach to determine HLs has been previously reported in our publications, such as Lotan et al. (2005, 2007) and Goler Baron et al. (2008). The new results are shown in Fig. S3B. They are consistent with our conclusion that Sfp1 stabilizes mRNAs.
Using a repressible promoter to determine mRNA HL is, unfortunately, not suitable in this paper because the promoter itself is involved in HL regulation. This observation is supported by Bregman et al. (2011) and depicted in Fig. 3, which illustrates that the promoter is critical for mRNA imprinting, consequently regulating HL.
(7) The effects of SFP1 on transcription were investigated by chromatin purification with Rpb3, a subunit of RNA polymerase, and the results were compared with synthesis rates determined by genomic run-on experiments. The decrease in polII presence on transcripts in the absence of SFP1 was not accompanied by a marked decrease in transcript output, suggesting an effect of Sfp1 in ensuring robust transcription and avoiding RNA polymerase backtracking. To further investigate the phenotypes associated with the depletion or absence of Sfp1, the authors examined the presence of Rpb4 along transcription units compared to Rpb3. One effect of spf1 deficiency was that this ratio, which decreased from the start of transcription towards the end of transcripts, increased slightly. The results presented are largely correlative and could arise from the focus on very specific types of mRNAs, such as those of ribosomal protein genes, which are sensitive to stress and are targeted by very active RNA degradation mechanisms activated, for example, under heat stress (Bresson et al., 2020).
Figure 7A illustrates a significant reduction in Rpb4/Rpb3 ratios along the transcription unit in WT cells. This reduction is notably more pronounced in CRAC+ genes compared to the highly transcribed quartile (Q1), which includes all ribosomal protein (RP) genes, and it is completely absent in sfp1∆ cells. Furthermore, it's important to highlight that the CRAC+ gene group displays a wide range of transcription rates, as measured by either Rpb3 ChIP or GRO (Figure 6A). Given these observations, we do not think that heightened sensitivity of RP mRNA degradation in response to stress is responsible for the pronounced difference in the configuration of the Pol II elongation complex that is detected in CRAC+ genes, mainly because this experiment was performed under standard (non-stress) culture conditions.
Correlative studies are particularly informative when a gene mutation eliminates a correlation, and this is precisely the type of study depicted in Figure 7B-C. The correlations shown in these panels are dependent on Sfp1. Indeed, RP genes are sensitive to stress. However, we used non-stressed conditions. Furthermore, CRAC+ genes did not display any apparent unusual destabilization but rather exhibited higher (not lower) mRNA stability compared to non-CRAC+ genes (Figure 7C).
-

-

Author Response
The authors wish to thank the Reviewers for valuable and constructive comments that will help up improve the paper’s quality.
Public Reviews:
Reviewer #1 (Public Review):
Summary:
This manuscript builds upon the authors' previous work on the cross-talk between transcription initiation and post-transcriptional events in yeast gene expression. These prior studies identified an mRNA 'imprinting' phenomenon linked to genes activated by the Rap1 transcription factor (TF), a surprising role for the Sfp1 TF in promoting RNA polymerase II (RNAPII) backtracking, and a role for the non-essential RNAPII subunits Rpb4/7 in the regulation of mRNA decay and translation. Here the authors aimed to extend these observations to provide a more coherent picture of the role of Sfp1 in transcription initiation and subsequent steps in …
Author Response
The authors wish to thank the Reviewers for valuable and constructive comments that will help up improve the paper’s quality.
Public Reviews:
Reviewer #1 (Public Review):
Summary:
This manuscript builds upon the authors' previous work on the cross-talk between transcription initiation and post-transcriptional events in yeast gene expression. These prior studies identified an mRNA 'imprinting' phenomenon linked to genes activated by the Rap1 transcription factor (TF), a surprising role for the Sfp1 TF in promoting RNA polymerase II (RNAPII) backtracking, and a role for the non-essential RNAPII subunits Rpb4/7 in the regulation of mRNA decay and translation. Here the authors aimed to extend these observations to provide a more coherent picture of the role of Sfp1 in transcription initiation and subsequent steps in gene expression. They provide evidence for (1) a physical interaction between Sfp1 and Rpb4, (2) Sfp1 binding and stabilization of mRNAs derived from genes whose promoters are bound by both Rap1 and Sfp1 and (3) an effect of Sfp1 on Rpb4 binding or conformation during transcription elongation.
Strengths:
This study provides evidence that a TF (yeast Sfp1), in addition to stimulating transcription initiation, can at some target genes interact with their mRNA transcripts and promote their stability. Sfp1 thus has a positive effect on two distinct regulatory steps. Furthermore, evidence is presented indicating that strong Sfp1 mRNA association requires both Rap1 and Sfp1 promoter binding and is increased at a sequence motif near the polyA track of many target mRNAs. Finally, they provide compelling evidence that Sfp1-bound mRNAs have higher levels of RNAPII backtracking and altered Rpb4 association or conformation compared to those not bound by Sfp1.
Weaknesses:
The Sfp1-Rpb4 association is supported only by a two-hybrid assay that is poorly described and lacks an important control. Furthermore, there is no evidence that this interaction is direct, nor are the interaction domains on either protein identified (or mutated to address function).
Indeed, our two hybrid, immunoprecipitation and imaging results do not allow us to conclusively discern whether the interaction between Rpb4 and Sfp1 is direct or indirect. While the interaction holds significance, we consider the direct versus indirect distinction to be of secondary importance in the context of this paper. We intend to give more attention to this matter in our revised paper. In addition, we will make an effort to investigate an in vitro interaction between Sfp1 and Rpb4 by employing purified Sfp1 and Rpb4 proteins.
The contention that Sfp1 nuclear export to the cytoplasm is transcription-dependent is not well supported by the experiments shown, which are not properly described in the text and are not accompanied by any primary data.
We note that this assay has been developed and published in prior research by Lee, M. S., M. Henry, and P. A. Silver. (G&D, 1996) and was reported in a number of subsequent papers. Reassuringly, our conclusion is supported by the observation that Sfp1 binds to Pol II transcripts co-transcriptionally suggesting that Sfp1 is exported in the context of the mRNA.
The presence of Sfp1 in P-bodies is of unclear relevance and the authors do not ask whether Sfp1-bound mRNAs are also present in these condensates.
In the revised paper, we will indicate that we do not know whether RP mRNAs are present in the actual foci shown in Fig. 1B.
Further analysis of Sfp1-bound mRNAs would be of interest, particularly to address the question of whether those from ribosomal protein genes and other growth-related genes that are known to display Sfp1 binding in their promoters are regulated (either stabilized or destabilized) by Sfp1.
Fig. 4A, C and D show that RP mRNAs become destabilized in sfp1Δ cells.
The authors need to discuss, and ideally address, the apparent paradox that their previous findings showed that Rap1 acts to destabilize its downstream transcripts, i.e. that it has the opposite effect of Sfp1 shown here.
We would like to thank Reviewer 1 for this valuable comment. In the revised paper, we will delve into our hypothesis suggesting that Rap1 is likely responsible for regulating the imprinting of other proteins, that, in turn, lead to the destabilization of mRNAs, such as Rpb4.
Finally, recent studies indicate that the drugs used here to measure mRNA stability induce a strong stress response accompanied by rapid and complex effects on transcription. Their relevance to mRNA stability in unstressed cells is questionable.
Half-lives were determined mainly by the GRO analysis of optimally proliferating cells. This method does not requires any drug or stressful treatment. The results obtained by this method were consistent with the those obtained after thiolutin addition. Nevertheless, in our revised manuscript, we plan to supplement the half-life data with results obtained by subjecting cells to a temperature shift to 42°C, a natural method to block transcription in wild-type (WT) cells. This approach to determine half-lives has been previously reported in our publications, such as Lotan et al. (2005, 2007) and Goler Baron et al. (2008). This may rule out effects of the drug on halfe-life.
Reviewer #2 (Public Review):
Summary:
The manuscript by Kelbert et al. presents results on the involvement of the yeast transcription factor Sfp1 in the stabilisation of transcripts whose synthesis it stimulates. Sfp1 is known to affect the synthesis of a number of important cellular transcripts, such as many of those that code for ribosomal proteins. The hypothesis that a transcription factor can remain bound to the nascent transcript and affect its cytoplasmic half-life is attractive, but the methods used to demonstrate the half-life effects and the association of Sfp1 with cytoplasmic transcripts remain to be fully validated, as explained in my comments on the results below:
Comments on methodology and results:
- A two-hybrid-based assay for protein-protein interactions identified Sfp1, a transcription factor known for its effects on ribosomal protein gene expression, as interacting with Rpb4, a subunit of RNA polymerase II. Classical two-hybrid experiments depend on the presence of the tested proteins in the nucleus of yeast cells, suggesting that the observed interaction occurs in the nucleus. Unfortunately, the two-hybrid method cannot determine whether the interaction is direct or mediated by nucleic acids.
Please see our response to comment 1 of Reviewer 1.
- Inactivation of nup49, a component of the nuclear pore complex, resulted in the redistribution of GFP-Sfp1 into the cytoplasm at the temperature non-permissive for the nup49-313 strain, suggesting that GFP-Sfp1 is a nucleo-cytoplasmic shuttling protein. This observation confirmed the dynamic nature of the nucleo-cytoplasmic distribution of Sfp1. For example, a similar redistribution to the cytoplasm was previously reported following rapamycin treatment and under starvation (Marion et al., PNAS 2004). In conjunction with the observation of an interaction with Rpb4, the authors observed slower nuclear import kinetics for GFP-Sfp1 in the absence of Rpb4 when cells were transferred to a glucose-containing medium after a period of starvation. Since the redistribution of GFP-Sfp1 was abolished in an rpb1-1/nup49-313 double mutant, the authors concluded that Sfp1 localisation to the cytoplasm depends on transcription. The double mutant yeast cells may show a variety of non-specific effects at the restrictive temperature, and whether transcription is required for Sfp1 cytoplasmic localisation remains incompletely demonstrated.
We concur with Reviewer 2 that any heat inactivation of a temperature-sensitive (ts) protein can result in non-specific effects. In the instance of rpb1-1, these non-specific effects are anticipated because of the transcriptional arrest, which can eventually lead to a reduction in protein content. However, it is worth noting that this process takes some time, whereas the impact on export is more rapid. We note that that this assay has been developed and published in prior research by Pam Silver (op. cit.) and was reported in a number of subsequent papers. Reassuringly, our conclusion is supported by the observation that Sfp1 binds to Pol II transcripts co-transcriptionally.
- Under starvation conditions, which led to the presence of Sfp1 in the cytoplasm and have previously been correlated with a decrease in the transcription of Sfp1 target genes, the authors observed that a plasmid-based expressed GFP-Sfp1 accumulated in cytoplasmic foci. These foci were also labelled by P-body markers such as Dcp2 and Lsm1. The quality of the microscopic images provided does not allow to determine whether Rpb4-RFP colocalises with GFP-Sfp1.
The submitted PDF figure is of low quality. We believe that high quality figure will be convincing.
- To understand to which RNA Sfp1 might bind, the authors used an N-terminally tagged fusion protein in a cross-linking and purification experiment. This method identified 264 transcripts for which the CRAC signal was considered positive and which mostly correspond to abundant mRNAs, including 74 ribosomal protein mRNAs or metabolic enzyme-abundant mRNAs such as PGK1. The authors did not provide evidence for the specificity of the observed CRAC signal, in particular, what would be the background of a similar experiment performed without UV cross-linking. In a validation experiment, the presence of several mRNAs in a purified SFP1 fraction was measured at levels that reflect the relative levels of RNA in a total RNA extract. Negative controls showing that abundant mRNAs not found in the CRAC experiment were clearly depleted from the purified fraction with Sfp1 would be crucial to assessing the specificity of the observed protein-RNA interactions. The CRAC-selected mRNAs were enriched for genes whose expression was previously shown to be upregulated upon Sfp1 overexpression (Albert et al., 2019). The presence of unspliced RPL30 pre-mRNA in the Sfp1 purification was interpreted as a sign of co-transcriptional assembly of Sfp1 into mRNA, but in the absence of valid negative controls, this hypothesis would require further experimental validation.
We argue that the 264 CRAC+ genes represent a distinct group with many unique features. Moreover, many CRAC+ genes do not fall into the category of highly transcribed genes.
The biological significance of the 264 CRAC+ mRNAs was demonstrated by various experiments; all are inconsistent with technical flaws. Some examples are:
Fig. 2a and B show that most reads of CRAC+ mRNA were mapped to specific location – close the pA sites.
Fig. 2C shows that most reads of CRAC+ mRNA were mapped to specific RNA motif.
Most RiBi CRAC+ promoter contain Rap1 binding sites (p= 1.9x10-22), whereas the vast majority of RiBi CRAC- promoters do not contain Rap1 binding site. (Fig. 3C).
Fig. 4A shows that RiBi CRAC+ mRNAs become destabilized due to Sfp1 deletion, whereas RiBi CRAC- mRNAs do not. Fig. 4B shows similar results due to
Fig. 6B shows that the impact of Sfp1 on backtracking is substantially higher for CRAC+ than for CRAC- genes. This is most clearly visible in RiBi genes.
Fig. 7A shows that the Sfp1-dependent changes along the transcription units is substantially more rigorous for CRAC+ than for CRAC-.
Fig. S4B Shows that chromatin binding profile of Sfp1 is different for CRAC+ and CRAC- genes
Moreover, only a portion of the RiBi mRNAs binds Sfp1, despite similar expression of all RiBi.
Most importantly, these genes do not all fall into the category of highly transcribed genes. On the contrary, as depicted in Figure 6A (green dots), it is evident that CRAC+ genes exhibit a diverse range of Rpb3 ChIP and GRO signals. Furthermore, as illustrated in Figure 7A, when comparing CRAC+ to Q1 (the most highly transcribed genes), it becomes evident that the Rpb4/Rpb3 profile of CRAC+ genes is not a result of high transcription levels. In our revised paper, we will give increased attention to this matter in the Discussion section.
- To address the important question of whether co-transcriptional assembly of Spf1 with transcripts could alter their stability, the authors first used a reporter system in which the RPL30 transcription unit is transferred to vectors under different transcriptional contexts, as previously described by the Choder laboratory (Bregman et al. 2011). While RPL30 expressed under an ACT1 promoter was barely detectable, the highest levels of RNA were observed in the context of the native upstream RPL30 sequence when Rap1 binding sites were also present. Sfp1 showed better association with reporter mRNAs containing Rap1 binding sites in the promoter region. However, removal of the Rap1 binding sites from the reporter vector also led to a drastic decrease in reporter mRNA levels. Whether the fraction of co-purified RNA is nuclear and co-transcriptional or not cannot be inferred from these results.
The proposed co-transcriptional binding of Sfp1 is based on the findings presented in Figure 5C and Figure S2D, as well as the observed binding of Sfp1 to transcripts containing introns, as shown in Figures 2D and 3B. Our conclusion, which we still uphold, was drawn from the results presented in Figure 3. These results led us to the assertion that the "RNA-binding capacity of Sfp1 is regulated by Rap1-binding sites located at the promoter." We maintain our stance on this conclusion. Indeed, the Rap1 binding site does impact mRNA levels, as highlighted by Reviewer 2. However, "construct E," which possesses a promoter with a Rap1 binding site, exhibits lower transcript levels compared to "construct F," which lacks such a binding site in its promoter. Despite this difference in transcript levels, Sfp1 was able to pull down the former transcript but not the latter, even though expression of the former gene is relatively low. Thus, the results appear to be more reliant on the specific capacity of Sfp1 to interact with the transcript rather than on the transcript's expression level.
- To complement the biochemical data presented in the first part of the manuscript, the authors turned to the deletion or rapid depletion of SFP1 and used labelling experiments to assess changes in the rate of synthesis, abundance, and decay of mRNAs under these conditions. An important observation was that in the absence of Sfp1, mRNAs encoding ribosomal protein genes not only had a reduced synthesis rate but also an increased degradation rate. This important observation needs careful validation, as genomic run-on experiments were used to measure half-lives, and this particular method was found to give results that correlated poorly with other measures of half-life in yeast (e.g. Chappelboim et al., 2022 for a comparison). Similarly, the use of thiolutin to block transcription as a method of assessing mRNA half-life has been reported to be problematic, as thiolutin can specifically inhibit the degradation of ribosomal protein mRNA (Pelechano & Perez-Ortin, 2008). Specific repressible reporters, such as those used by Baudrimont et al. (2017), would need to be tested to validate the effect of Sfp1 on the half-life of specific mRNAs. Also, it would be very difficult to infer from the images presented whether the rate of deadenylation is altered by Sfp1.
Various methods exist for assessing mRNA half-lives (HLs), and each of them carries its own set of challenges and biases. Consequently, it becomes problematic to directly compare HL values of a specific mRNA when different methods are employed. The superiority of one particular method over others remains unclear. However, they all exhibit a high degree of reliability when it comes to comparing different strains under the identical conditions using a single method.
Estimating half-lives through the GRO approach is a non-invasive method, applied on optimally proliferating cells, which has been employed in numerous publications. While no method is without its limitations, we consider this approach to be among the most dependable. Our HL determination using thiolutin to block transcription provided results that were consistent with the values obtained by the GRO approach.
Nevertheless, in our revised manuscript, we plan to supplement the HL data, obtain by thiolutin, with results obtained by subjecting cells to a temperature shift to 42°C, a natural method to block transcription in wild-type (WT) cells. This approach to determine HLs has been previously reported in our publications, such as Lotan et al. (2005, 2007) and Goler Baron et al. (2008).
- The effects of SFP1 on transcription were investigated by chromatin purification with Rpb3, a subunit of RNA polymerase, and the results were compared with synthesis rates determined by genomic run-on experiments. The decrease in polII presence on transcripts in the absence of SFP1 was not accompanied by a marked decrease in transcript output, suggesting an effect of Sfp1 in ensuring robust transcription and avoiding RNA polymerase backtracking. To further investigate the phenotypes associated with the depletion or absence of Sfp1, the authors examined the presence of Rpb4 along transcription units compared to Rpb3. One effect of spf1 deficiency was that this ratio, which decreased from the start of transcription towards the end of transcripts, increased slightly. The results presented are largely correlative and could arise from the focus on very specific types of mRNAs, such as those of ribosomal protein genes, which are sensitive to stress and are targeted by very active RNA degradation mechanisms activated, for example, under heat stress (Bresson et al., 2020).
Figure 7A illustrates a significant reduction in Rpb4/Rpb3 ratios along the transcription unit in WT cells. This reduction is notably more pronounced in CRAC+ genes compared to the highly transcribed quartile (Q1), which includes all ribosomal protein (RP) genes, and it is completely absent in sfp1∆ cells. Furthermore, it's important to highlight that the CRAC+ gene group displays a wide range of transcription rates, as measured by either Rpb3 ChIP or GRO (Figure 6A). Given these observations, it is challenging to reconcile how the heightened sensitivity of RP mRNA degradation in response to stress could account for the more pronounced differences in the configuration of the Pol II elongation complex that are detected in CRAC+ genes under standard culture conditions in wt cells.
Correlative studies are particularly informative when a gene mutation eliminates a correlation, and this is precisely the type of study depicted in Figure 7B-C. The configuration of elongating Pol II (as reflected by Rpb4/Rpb3 ratios) and the backtracking index are both transcriptional outputs. It is difficult to envision how stress-induced destabilization of RP mRNAs could explain the twofold higher correlation between these two parameters observed in CRAC+ genes under non-stressful conditions in WT cells (Figure 7B).
Furthermore, it's worth noting that in WT cells, CRAC+ genes did not display any apparent unusual destabilization, but rather exhibited higher (not lower) mRNA stability compared to CRAC- genes (Figure 7C).
Strengths:
- Diversity of experimental approaches used
- Validation of large-scale results with appropriate reporters
Weaknesses:
- Choice of evaluation method to test mRNA half-life
- Lack of controls for the CRAC results
-

eLife assessment
This study shows that the yeast transcription factor Sfp1 binds to a subset of its target gene mRNAs, increases their half-lives, and affects RNA polymerase II backtracking. These, and other related findings, provide important new insights into mechanisms by which a transcription factor can affect post-transcriptional steps in gene regulation. The main claims are partially backed by the evidence presented. However, the evidence remains incomplete as the methods used to estimate RNA degradation rates and the biochemistry of Sfp1-RNA complexes require further validation.
-

Reviewer #1 (Public Review):
Summary:
This manuscript builds upon the authors' previous work on the cross-talk between transcription initiation and post-transcriptional events in yeast gene expression. These prior studies identified an mRNA 'imprinting' phenomenon linked to genes activated by the Rap1 transcription factor (TF), a surprising role for the Sfp1 TF in promoting RNA polymerase II (RNAPII) backtracking, and a role for the non-essential RNAPII subunits Rpb4/7 in the regulation of mRNA decay and translation. Here the authors aimed to extend these observations to provide a more coherent picture of the role of Sfp1 in transcription initiation and subsequent steps in gene expression. They provide evidence for (1) a physical interaction between Sfp1 and Rpb4, (2) Sfp1 binding and stabilization of mRNAs derived from genes whose …Reviewer #1 (Public Review):
Summary:
This manuscript builds upon the authors' previous work on the cross-talk between transcription initiation and post-transcriptional events in yeast gene expression. These prior studies identified an mRNA 'imprinting' phenomenon linked to genes activated by the Rap1 transcription factor (TF), a surprising role for the Sfp1 TF in promoting RNA polymerase II (RNAPII) backtracking, and a role for the non-essential RNAPII subunits Rpb4/7 in the regulation of mRNA decay and translation. Here the authors aimed to extend these observations to provide a more coherent picture of the role of Sfp1 in transcription initiation and subsequent steps in gene expression. They provide evidence for (1) a physical interaction between Sfp1 and Rpb4, (2) Sfp1 binding and stabilization of mRNAs derived from genes whose promoters are bound by both Rap1 and Sfp1 and (3) an effect of Sfp1 on Rpb4 binding or conformation during transcription elongation.Strengths:
This study provides evidence that a TF (yeast Sfp1), in addition to stimulating transcription initiation, can at some target genes interact with their mRNA transcripts and promote their stability. Sfp1 thus has a positive effect on two distinct regulatory steps. Furthermore, evidence is presented indicating that strong Sfp1 mRNA association requires both Rap1 and Sfp1 promoter binding and is increased at a sequence motif near the polyA track of many target mRNAs. Finally, they provide compelling evidence that Sfp1-bound mRNAs have higher levels of RNAPII backtracking and altered Rpb4 association or conformation compared to those not bound by Sfp1.Weaknesses:
The Sfp1-Rpb4 association is supported only by a two-hybrid assay that is poorly described and lacks an important control. Furthermore, there is no evidence that this interaction is direct, nor are the interaction domains on either protein identified (or mutated to address function).The contention that Sfp1 nuclear export to the cytoplasm is transcription-dependent is not well supported by the experiments shown, which are not properly described in the text and are not accompanied by any primary data.
The presence of Sfp1 in P-bodies is of unclear relevance and the authors do not ask whether Sfp1-bound mRNAs are also present in these condensates.Further analysis of Sfp1-bound mRNAs would be of interest, particularly to address the question of whether those from ribosomal protein genes and other growth-related genes that are known to display Sfp1 binding in their promoters are regulated (either stabilized or destabilized) by Sfp1.
The authors need to discuss, and ideally address, the apparent paradox that their previous findings showed that Rap1 acts to destabilize its downstream transcripts, i.e. that it has the opposite effect of Sfp1 shown here.
Finally, recent studies indicate that the drugs used here to measure mRNA stability induce a strong stress response accompanied by rapid and complex effects on transcription. Their relevance to mRNA stability in unstressed cells is questionable.
-

Reviewer #2 (Public Review):
Summary:
The manuscript by Kelbert et al. presents results on the involvement of the yeast transcription factor Sfp1 in the stabilisation of transcripts whose synthesis it stimulates. Sfp1 is known to affect the synthesis of a number of important cellular transcripts, such as many of those that code for ribosomal proteins. The hypothesis that a transcription factor can remain bound to the nascent transcript and affect its cytoplasmic half-life is attractive, but the methods used to demonstrate the half-life effects and the association of Sfp1 with cytoplasmic transcripts remain to be fully validated, as explained in my comments on the results below:Comments on methodology and results:
(1) A two-hybrid-based assay for protein-protein interactions identified Sfp1, a transcription factor known for its effects …Reviewer #2 (Public Review):
Summary:
The manuscript by Kelbert et al. presents results on the involvement of the yeast transcription factor Sfp1 in the stabilisation of transcripts whose synthesis it stimulates. Sfp1 is known to affect the synthesis of a number of important cellular transcripts, such as many of those that code for ribosomal proteins. The hypothesis that a transcription factor can remain bound to the nascent transcript and affect its cytoplasmic half-life is attractive, but the methods used to demonstrate the half-life effects and the association of Sfp1 with cytoplasmic transcripts remain to be fully validated, as explained in my comments on the results below:Comments on methodology and results:
(1) A two-hybrid-based assay for protein-protein interactions identified Sfp1, a transcription factor known for its effects on ribosomal protein gene expression, as interacting with Rpb4, a subunit of RNA polymerase II. Classical two-hybrid experiments depend on the presence of the tested proteins in the nucleus of yeast cells, suggesting that the observed interaction occurs in the nucleus. Unfortunately, the two-hybrid method cannot determine whether the interaction is direct or mediated by nucleic acids.(2) Inactivation of nup49, a component of the nuclear pore complex, resulted in the redistribution of GFP-Sfp1 into the cytoplasm at the temperature non-permissive for the nup49-313 strain, suggesting that GFP-Sfp1 is a nucleo-cytoplasmic shuttling protein. This observation confirmed the dynamic nature of the nucleo-cytoplasmic distribution of Sfp1. For example, a similar redistribution to the cytoplasm was previously reported following rapamycin treatment and under starvation (Marion et al., PNAS 2004). In conjunction with the observation of an interaction with Rpb4, the authors observed slower nuclear import kinetics for GFP-Sfp1 in the absence of Rpb4 when cells were transferred to a glucose-containing medium after a period of starvation. Since the redistribution of GFP-Sfp1 was abolished in an rpb1-1/nup49-313 double mutant, the authors concluded that Sfp1 localisation to the cytoplasm depends on transcription. The double mutant yeast cells may show a variety of non-specific effects at the restrictive temperature, and whether transcription is required for Sfp1 cytoplasmic localisation remains incompletely demonstrated.
(3) Under starvation conditions, which led to the presence of Sfp1 in the cytoplasm and have previously been correlated with a decrease in the transcription of Sfp1 target genes, the authors observed that a plasmid-based expressed GFP-Sfp1 accumulated in cytoplasmic foci. These foci were also labelled by P-body markers such as Dcp2 and Lsm1. The quality of the microscopic images provided does not allow to determine whether Rpb4-RFP colocalises with GFP-Sfp1.
(4) To understand to which RNA Sfp1 might bind, the authors used an N-terminally tagged fusion protein in a cross-linking and purification experiment. This method identified 264 transcripts for which the CRAC signal was considered positive and which mostly correspond to abundant mRNAs, including 74 ribosomal protein mRNAs or metabolic enzyme-abundant mRNAs such as PGK1. The authors did not provide evidence for the specificity of the observed CRAC signal, in particular, what would be the background of a similar experiment performed without UV cross-linking. In a validation experiment, the presence of several mRNAs in a purified SFP1 fraction was measured at levels that reflect the relative levels of RNA in a total RNA extract. Negative controls showing that abundant mRNAs not found in the CRAC experiment were clearly depleted from the purified fraction with Sfp1 would be crucial to assessing the specificity of the observed protein-RNA interactions. The CRAC-selected mRNAs were enriched for genes whose expression was previously shown to be upregulated upon Sfp1 overexpression (Albert et al., 2019). The presence of unspliced RPL30 pre-mRNA in the Sfp1 purification was interpreted as a sign of co-transcriptional assembly of Sfp1 into mRNA, but in the absence of valid negative controls, this hypothesis would require further experimental validation.
(5) To address the important question of whether co-transcriptional assembly of Spf1 with transcripts could alter their stability, the authors first used a reporter system in which the RPL30 transcription unit is transferred to vectors under different transcriptional contexts, as previously described by the Choder laboratory (Bregman et al. 2011). While RPL30 expressed under an ACT1 promoter was barely detectable, the highest levels of RNA were observed in the context of the native upstream RPL30 sequence when Rap1 binding sites were also present. Sfp1 showed better association with reporter mRNAs containing Rap1 binding sites in the promoter region. However, removal of the Rap1 binding sites from the reporter vector also led to a drastic decrease in reporter mRNA levels. Whether the fraction of co-purified RNA is nuclear and co-transcriptional or not cannot be inferred from these results.
(6) To complement the biochemical data presented in the first part of the manuscript, the authors turned to the deletion or rapid depletion of SFP1 and used labelling experiments to assess changes in the rate of synthesis, abundance, and decay of mRNAs under these conditions. An important observation was that in the absence of Sfp1, mRNAs encoding ribosomal protein genes not only had a reduced synthesis rate but also an increased degradation rate. This important observation needs careful validation, as genomic run-on experiments were used to measure half-lives, and this particular method was found to give results that correlated poorly with other measures of half-life in yeast (e.g. Chappelboim et al., 2022 for a comparison). Similarly, the use of thiolutin to block transcription as a method of assessing mRNA half-life has been reported to be problematic, as thiolutin can specifically inhibit the degradation of ribosomal protein mRNA (Pelechano & Perez-Ortin, 2008). Specific repressible reporters, such as those used by Baudrimont et al. (2017), would need to be tested to validate the effect of Sfp1 on the half-life of specific mRNAs. Also, it would be very difficult to infer from the images presented whether the rate of deadenylation is altered by Sfp1.
(7) The effects of SFP1 on transcription were investigated by chromatin purification with Rpb3, a subunit of RNA polymerase, and the results were compared with synthesis rates determined by genomic run-on experiments. The decrease in polII presence on transcripts in the absence of SFP1 was not accompanied by a marked decrease in transcript output, suggesting an effect of Sfp1 in ensuring robust transcription and avoiding RNA polymerase backtracking. To further investigate the phenotypes associated with the depletion or absence of Sfp1, the authors examined the presence of Rpb4 along transcription units compared to Rpb3. One effect of spf1 deficiency was that this ratio, which decreased from the start of transcription towards the end of transcripts, increased slightly. The results presented are largely correlative and could arise from the focus on very specific types of mRNAs, such as those of ribosomal protein genes, which are sensitive to stress and are targeted by very active RNA degradation mechanisms activated, for example, under heat stress (Bresson et al., 2020).
Strengths:
- Diversity of experimental approaches used
- Validation of large-scale results with appropriate reportersWeaknesses:
- Choice of evaluation method to test mRNA half-life
- Lack of controls for the CRAC results -