Positive selection and relaxed purifying selection contribute to rapid evolution of male-biased genes in a dioecious flowering plant
Curation statements for this article:-
Curated by eLife
eLife assessment
This valuable paper examines gene expression differences between male and female individuals over the course of flower development in the dioecious angiosperm Trichosantes pilosa. Male-biased genes evolve faster than female-biased and unbiased genes, which is frequently observed in animals, but this is the first report of such a pattern in plants. In spite of the limited sample size, the evidence is mostly solid and the methods appropriate for a non-model organism. The resources produced will be used by researchers working in the Cucurbitaceae, and the results obtained advance our understanding of the mechanisms of plant sexual reproduction and its evolutionary implications: as such they will broadly appeal to evolutionary biologists and plant biologists.
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Sex-biased genes offer insights into the evolution of sexual dimorphism. Sex-biased genes, especially those with male bias, show elevated evolutionary rates of protein sequences driven by positive selection and relaxed purifying selection in animals. Although rapid sequence evolution of sex-biased genes and evolutionary forces have been investigated in animals and brown algae, less is known about evolutionary forces in dioecious angiosperms. In this study, we separately compared the expression of sex-biased genes between female and male floral buds and between female and male flowers at anthesis in dioecious Trichosanthes pilosa (Cucurbitaceae). In floral buds, sex-biased gene expression was pervasive, and had significantly different roles in sexual dimorphism such as physiology. We observed higher rates of sequence evolution for male-biased genes in floral buds compared to female-biased and unbiased genes. Male-biased genes under positive selection were mainly associated with functions to abiotic stress and immune responses, suggesting that high evolutionary rates are driven by adaptive evolution. Additionally, relaxed purifying selection may contribute to accelerated evolution in male-biased genes generated by gene duplication. Our findings, for the first time in angiosperms, suggest evident rapid evolution of male-biased genes, advance our understanding of the patterns and forces driving the evolution of sexual dimorphism in dioecious plants.
Article activity feed
-

-

-

-

Author Response
The following is the authors’ response to the previous reviews.
eLife assessment
This valuable paper examines gene expression differences between male and female individuals over the course of flower development in the dioecious angiosperm Trichosantes pilosa. Male-biased genes evolve faster than female-biased and unbiased genes, which is frequently observed in animals, but this is the first report of such a pattern in plants. In spite of the limited sample size, the evidence is mostly solid and the methods appropriate for a non-model organism. The resources produced will be used by researchers working in the Cucurbitaceae, and the results obtained advance our understanding of the mechanisms of plant sexual reproduction and its evolutionary implications: as such they will broadly appeal to evolutionary biologists and …
Author Response
The following is the authors’ response to the previous reviews.
eLife assessment
This valuable paper examines gene expression differences between male and female individuals over the course of flower development in the dioecious angiosperm Trichosantes pilosa. Male-biased genes evolve faster than female-biased and unbiased genes, which is frequently observed in animals, but this is the first report of such a pattern in plants. In spite of the limited sample size, the evidence is mostly solid and the methods appropriate for a non-model organism. The resources produced will be used by researchers working in the Cucurbitaceae, and the results obtained advance our understanding of the mechanisms of plant sexual reproduction and its evolutionary implications: as such they will broadly appeal to evolutionary biologists and plant biologists.
Public Reviews:
Reviewer #1 (Public Review):
The evolution of dioecy in angiosperms has significant implications for plant reproductive efficiency, adaptation, evolutionary potential, and resilience to environmental changes. Dioecy allows for the specialization and division of labor between male and female plants, where each sex can focus on specific aspects of reproduction and allocate resources accordingly. This division of labor creates an opportunity for sexual selection to act and can drive the evolution of sexual dimorphism.
In the present study, the authors investigate sex-biased gene expression patterns in juvenile and mature dioecious flowers to gain insights into the molecular basis of sexual dimorphism. They find that a large proportion of the plant transcriptome is differentially regulated between males and females with the number of sex-biased genes in floral buds being approximately 15 times higher than in mature flowers. The functional analysis of sex-biased genes reveals that chemical defense pathways against herbivores are up-regulated in the female buds along with genes involved in the acquisition of resources such as carbon for fruit and seed production, whereas male buds are enriched in genes related to signaling, inflorescence development and senescence of male flowers. Furthermore, the authors implement sophisticated maximum likelihood methods to understand the forces driving the evolution of sex-biased genes. They highlight the influence of positive and relaxed purifying selection on the evolution of male-biased genes, which show significantly higher rates of non-synonymous to synonymous substitutions than female or unbiased genes. This is the first report (to my knowledge) highlighting the occurrence of this pattern in plants. Overall, this study provides important insights into the genetic basis of sexual dimorphism and the evolution of reproductive genes in Cucurbitaceae.
Reviewer #2 (Public Review):
Summary:
This study uses transcriptome sequence from a dioecious plant to compare evolutionary rates between genes with male- and female-biased expression and distinguish between relaxed selection and positive selection as causes for more rapid evolution. These questions have been explored in animals and algae, but few studies have investigated this in dioecious angiosperms, and none have so far identified faster rates of evolution in male-biased genes (though see Hough et al. 2014 https://doi.org/10.1073/pnas.1319227111).
Strengths:
The methods are appropriate to the questions asked. Both the sample size and the depth of sequencing are sufficient, and the methods used to estimate evolutionary rates and the strength of selection are appropriate. The data presented are consistent with faster evolution of genes with male-biased expression, due to both positive and relaxed selection.
This is a useful contribution to understanding the effect of sex-biased expression in genetic evolution in plants. It demonstrates the range of variation in evolutionary rates and selective mechanisms, and provides further context to connect these patterns to potential explanatory factors in plant diversity such as the age of sex chromosomes and the developmental trajectories of male and female flowers.
Weaknesses:
The presence of sex chromosomes is a potential confounding factor, since there are different evolutionary expectations for X-linked, Y-linked, and autosomal genes. Attempting to distinguish transcripts on the sex chromosomes from autosomal transcripts could provide additional insight into the relative contributions of positive and relaxed selection.
Reviewer #3 (Public Review):
The potential for sexual selection and the extent of sexual dimorphism in gene expression have been studied in great detail in animals, but hardly examined in plants so far. In this context, the study by Zhao, Zhou et al. al represents a welcome addition to the literature.
Relative to the previous studies in Angiosperms, the dataset is interesting in that it focuses on reproductive rather than somatic tissues (which makes sense to investigate sexual selection), and includes more than a single developmental stage (buds + mature flowers).
Recommendations for the authors:
Reviewer #3 (Recommendations For The Authors):
I have reviewed this new version and find that it now addresses some of the shortcomings of the previous manuscript. However, several important limitations still remain:
- The conclusion that sex-linked genes contribute relatively little to the patterns described is important and would be worth including in the manuscript briefly (not just the response letter), focusing for instance on the overall comparable proportions of sex-linked genes among male-biased (3/343=0.087%), female-biased (19/1145=1.66%) and unbiased genes (36/2378=1.51%).
Authors’ response: Thank you for your advice. We have added these sentences in “Discussion” section (Lines 492-499).
- The new sentence included in the results "we also found that most of them were members of different gene families generated by gene duplication" is too vague. The motivation of this analysis is not explained, leaving the intended message unclear.
Authors’ response: In the previous revision, as stressed by reviewer #1 “(2) Paragraph (407-416) describes the analysis of duplicated genes under relaxed selection but there is no mention of this in the results”, we added the sentence “we also found that most of them were members of different gene families generated by gene duplication” in “Relaxed selection” paragraph of the results. Accordingly, in “Discussion” section, we discussed the associations between gene duplication and relaxed selection (Lines 461-473).
Following your suggestion, we revised the results (Lines 304-307) to “Using the RELAX model, we detected that 18 out of 343 OGs (5.23%) showed significant evidence of relaxed selection (K = 0.0184–0.6497) (Tables S9). Most of the 18 OGs are members of different gene families generated by gene duplication (Table S13)”. This makes it more coherent with the discussion.
- The sentences "given that dN/dS values of sex-biased genes were higher due to codon usage bias..." are very confusing. I do not understand the argument being made here. I do not see why "lower dS rates would be expected in sex-biased genes ..."
Authors’ response: We respectfully argue that codon usage bias was positively related to synonymous substitution rates. That is, stronger codon usage bias may be related to higher synonymous substitution rates (Parvathy et al., 2022). Lower ENC values represent stronger codon usage bias. So, if ω (dN/dS) values of sex-biased genes are higher due to codon usage bias, we expect lower dS rates (That is, higher ENC values). Please refer to the relevant papers (e. g. Darolti et al., 2018; Catalan et al., 2018; Schrader et al., 2021, cited in the references of the paper).
- The manuscript now reports the proportion of unitigs annotated by similarity with a number of species. While this is an interesting observation, the reviewer was actually asking for a comparison between the number of unitigs (59,051) and the number of genes annotated in a typical cucurbitaceae genome. This would give an indication of the level of redundancy of the de novo assembled transcriptome.
Authors’ response: We admit that in the final assembly, transcripts may be overestimated. We respectfully suggest that it may be inappropriate to assess the redundancy of the de novo assembled transcriptome by comparing the transcriptome sequences with the genomic sequences. An appropriate approach is to compare transcriptome sequences and transcriptome sequences among different species. For example, Hu et al., 2020 (reference cited in the paper) obtained 145,975 non-redundant unigenes from flower buds of female and male plants in Trichosanthes kirilowii. Mohanty et al. (2017) obtained 71,823 non-redundant unigenes from flower buds of female and male plants in Coccinia grandis.
Reference:
Mohanty JN, Nayak S, Jha S, Joshi RK. 2017. Transcriptome profiling of the floral buds and discovery of genes related to sex-differentiation in the dioecious cucurbit Coccinia grandis (L.) Voigt. Gene. 626: 395-406.
- From reading the text I could not understand the extent to which the permutation test actually agreed with the Wilcoxon rank sum test. The text says that the results were "almost consistent", which is too vague. This paragraph should be clarified.
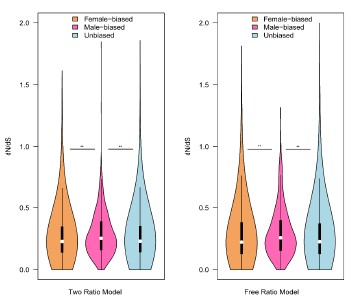
Authors’ response: We performed permutation test for sex-biased genes in floral buds and flowers at anthesis. However, only in floral buds, the results of both tests (permutation test and Wilcoxon rank sum test) are significant. Taking your suggestions in consideration, we have revised them as “Additionally, we found that only in floral buds, there were significant differences in ω values in the results of ‘free-ratio’ model (female-biased versus male-biased genes, P = 0.04282 and male-biased versus unbiased genes, P = 0.01114) and ‘two-ratio’ model (female-biased versus male-biased genes, P = 0.01992 and male-biased versus unbiased genes, P = 0.02127, respectively) by permutation t test, which is consistent with the results of Wilcoxon rank sum test.(Lines 273-280)”.
- The paragraph on the link between codon usage and dN/dS is very unclear and quite unnecessary. I would suggest to simply remove lines 312-323.
Authors’ response: We respectfully argue that codon usage bias is one of the most important factors for higher rates of sequence evolution. Please refer to Darolti et al. (2018), Catalan et al. (2018) and Schrader et al. (2021) (cited in the references of the paper). We retain these lines here.
- The discussion contains many unnecessary repeats from the introduction and results section. I suggest shortening drastically at several places, including:
- remove lines 367-369
Authors’ response: Thank you for your suggestion. We revised these lines to “In this study, we compared the expression profiles of sex-biased genes between sexes and two tissue types, investigated whether sex-biased genes exhibited evidence of rapid evolutionary rates of protein sequences and identified the evolutionary forces responsible for the observed patterns in the dioecious Trichosanthes pilosa (Lines 369-373)”.
We removed the sentence “We compared the expression profiles of sex-biased genes between sexes and two tissue types and examined the signatures of rapid sequence evolution for sex-biased genes, as well as the contributions of potential evolutionary forces. (Lines 374-376)”.
- remove lines 395-410
Authors’ response: Here we mainly discussed the possible associations between sex-biased genes, adaptation and sexual dimorphic traits. We retain them here for clarity.
- remove lines 449-483, as they are almost entirely repetitions of elements already made clear in the results section.
Authors’ response: In these paragraphs, we discussed reasons that lead to relaxed purifying selection for sex-biased genes. They are coherent with the results section. We retain them to make it clearer.
Minor comments:
- line 146: remove "However"
Authors’ response: We have revised it.
- line 187: "female flower buds tend to masculinize": the meaning is obscure
Authors’ response: We revised them as “Using hierarchical clustering analysis, we evaluated different levels of gene expression across sexes and tissues (Fig. 2C). Gene expression for female floral buds clustered most distantly from expression in female flowers at anthesis. However, expression in male floral buds clustered with expression in female flowers at anthesis, suggesting that male floral buds maybe tend to feminization in the early stages of floral development.”.
- line 226: "we sequenced transcriptomes of T. pilosa": rather say "we used the transcriptomes described above for T. pilosa"
Authors’ response: We have revised it.
- line 279: the meaning of "branch-site model A and branch site model null" is still not made clear.
Authors’ response: We have revised it.
- line 324: change to: "we also analysed whether female-biased and unbiased genes underwent... "
Authors’ response: We have revised it.
-

eLife assessment
This valuable paper examines gene expression differences between male and female individuals over the course of flower development in the dioecious angiosperm Trichosantes pilosa. Male-biased genes evolve faster than female-biased and unbiased genes, which is frequently observed in animals, but this is the first report of such a pattern in plants. In spite of the limited sample size, the evidence is mostly solid and the methods appropriate for a non-model organism. The resources produced will be used by researchers working in the Cucurbitaceae, and the results obtained advance our understanding of the mechanisms of plant sexual reproduction and its evolutionary implications: as such they will broadly appeal to evolutionary biologists and plant biologists.
-

Reviewer #1 (Public Review):
The evolution of dioecy in angiosperms has significant implications for plant reproductive efficiency, adaptation, evolutionary potential, and resilience to environmental changes. Dioecy allows for the specialization and division of labor between male and female plants, where each sex can focus on specific aspects of reproduction and allocate resources accordingly. This division of labor creates an opportunity for sexual selection to act and can drive the evolution of sexual dimorphism.
In the present study, the authors investigate sex-biased gene expression patterns in juvenile and mature dioecious flowers to gain insights into the molecular basis of sexual dimorphism. They find that a large proportion of the plant transcriptome is differentially regulated between males and females with the number of …
Reviewer #1 (Public Review):
The evolution of dioecy in angiosperms has significant implications for plant reproductive efficiency, adaptation, evolutionary potential, and resilience to environmental changes. Dioecy allows for the specialization and division of labor between male and female plants, where each sex can focus on specific aspects of reproduction and allocate resources accordingly. This division of labor creates an opportunity for sexual selection to act and can drive the evolution of sexual dimorphism.
In the present study, the authors investigate sex-biased gene expression patterns in juvenile and mature dioecious flowers to gain insights into the molecular basis of sexual dimorphism. They find that a large proportion of the plant transcriptome is differentially regulated between males and females with the number of sex-biased genes in floral buds being approximately 15 times higher than in mature flowers. The functional analysis of sex-biased genes reveals that chemical defense pathways against herbivores are up-regulated in the female buds along with genes involved in the acquisition of resources such as carbon for fruit and seed production, whereas male buds are enriched in genes related to signaling, inflorescence development and senescence of male flowers. Furthermore, the authors implement sophisticated maximum likelihood methods to understand the forces driving the evolution of sex-biased genes. They highlight the influence of positive and relaxed purifying selection on the evolution of male-biased genes, which show significantly higher rates of non-synonymous to synonymous substitutions than female or unbiased genes. This is the first report (to my knowledge) highlighting the occurrence of this pattern in plants. Overall, this study provides important insights into the genetic basis of sexual dimorphism and the evolution of reproductive genes in Cucurbitaceae.
-

Reviewer #2 (Public Review):
Summary:
This study uses transcriptome sequence from a dioecious plant to compare evolutionary rates between genes with male- and female-biased expression and distinguish between relaxed selection and positive selection as causes for more rapid evolution. These questions have been explored in animals and algae, but few studies have investigated this in dioecious angiosperms, and none have so far identified faster rates of evolution in male-biased genes (though see Hough et al. 2014 https://doi.org/10.1073/pnas.1319227111).
Strengths:
The methods are appropriate to the questions asked. Both the sample size and the depth of sequencing are sufficient, and the methods used to estimate evolutionary rates and the strength of selection are appropriate. The data presented are consistent with faster evolution of …
Reviewer #2 (Public Review):
Summary:
This study uses transcriptome sequence from a dioecious plant to compare evolutionary rates between genes with male- and female-biased expression and distinguish between relaxed selection and positive selection as causes for more rapid evolution. These questions have been explored in animals and algae, but few studies have investigated this in dioecious angiosperms, and none have so far identified faster rates of evolution in male-biased genes (though see Hough et al. 2014 https://doi.org/10.1073/pnas.1319227111).
Strengths:
The methods are appropriate to the questions asked. Both the sample size and the depth of sequencing are sufficient, and the methods used to estimate evolutionary rates and the strength of selection are appropriate. The data presented are consistent with faster evolution of genes with male-biased expression, due to both positive and relaxed selection.
This is a useful contribution to understanding the effect of sex-biased expression in genetic evolution in plants. It demonstrates the range of variation in evolutionary rates and selective mechanisms, and provides further context to connect these patterns to potential explanatory factors in plant diversity such as the age of sex chromosomes and the developmental trajectories of male and female flowers.
Weaknesses:
The presence of sex chromosomes is a potential confounding factor, since there are different evolutionary expectations for X-linked, Y-linked, and autosomal genes. Attempting to distinguish transcripts on the sex chromosomes from autosomal transcripts could provide additional insight into the relative contributions of positive and relaxed selection.
-

Reviewer #3 (Public Review):
The potential for sexual selection and the extent of sexual dimorphism in gene expression have been studied in great detail in animals, but hardly examined in plants so far. In this context, the study by Zhao, Zhou et al. al represents a welcome addition to the literature.
Relative to the previous studies in Angiosperms, the dataset is interesting in that it focuses on reproductive rather than somatic tissues (which makes sense to investigate sexual selection), and includes more than a single developmental stage (buds + mature flowers).
-

-

-

-

-

Author Response
The following is the authors’ response to the previous reviews.
eLife assessment
This valuable paper examines gene expression differences between male and female individuals over the course of flower development in the dioecious angiosperm Trichosantes pilosa. The authors show that male-biased genes evolve faster than female-biased and unbiased genes. This is frequently observed in animals, but this is the first report of such a pattern in plants. In spite of the limited sample size, the evidence is mostly solid and the methods appropriate for a non-model organism. The resources produced will be used by researchers working in the Cucurbitaceae, and the results obtained advance our understanding of the mechanisms of plant sexual reproduction and its evolutionary implications: as such they will broadly appeal to …
Author Response
The following is the authors’ response to the previous reviews.
eLife assessment
This valuable paper examines gene expression differences between male and female individuals over the course of flower development in the dioecious angiosperm Trichosantes pilosa. The authors show that male-biased genes evolve faster than female-biased and unbiased genes. This is frequently observed in animals, but this is the first report of such a pattern in plants. In spite of the limited sample size, the evidence is mostly solid and the methods appropriate for a non-model organism. The resources produced will be used by researchers working in the Cucurbitaceae, and the results obtained advance our understanding of the mechanisms of plant sexual reproduction and its evolutionary implications: as such they will broadly appeal to evolutionary biologists and plant biologists.
Public Reviews:
Reviewer #1 (Public Review):
The evolution of dioecy in angiosperms has significant implications for plant reproductive efficiency, adaptation, evolutionary potential, and resilience to environmental changes. Dioecy allows for the specialization and division of labor between male and female plants, where each sex can focus on specific aspects of reproduction and allocate resources accordingly. This division of labor creates an opportunity for sexual selection to act and can drive the evolution of sexual dimorphism.
In the present study, the authors investigate sex-biased gene expression patterns in juvenile and mature dioecious flowers to gain insights into the molecular basis of sexual dimorphism. They find that a large proportion of the plant transcriptome is differentially regulated between males and females with the number of sex-biased genes in floral buds being approximately 15 times higher than in mature flowers. The functional analysis of sex-biased genes reveals that chemical defense pathways against herbivores are up-regulated in the female buds along with genes involved in the acquisition of resources such as carbon for fruit and seed production, whereas male buds are enriched in genes related to signaling, inflorescence development and senescence of male flowers. Furthermore, the authors implement sophisticated maximum likelihood methods to understand the forces driving the evolution of sex-biased genes. They highlight the influence of positive and relaxed purifying selection on the evolution of male-biased genes, which show significantly higher rates of non-synonymous to synonymous substitutions than female or unbiased genes. This is the first report (to my knowledge) highlighting the occurrence of this pattern in plants. Overall, this study provides important insights into the genetic basis of sexual dimorphism and the evolution of reproductive genes in Cucurbitaceae.
Reviewer #2 (Public Review):
Summary:
This study uses transcriptome sequence from a dioecious plant to compare evolutionary rates between genes with male- and female-biased expression and distinguish between relaxed selection and positive selection as causes for more rapid evolution. These questions have been explored in animals and algae, but few studies have investigated this in dioecious angiosperms, and none have so far identified faster rates of evolution in male-biased genes (though see Hough et al. 2014 https://doi.org/10.1073/pnas.1319227111).
Strengths:
The methods are appropriate to the questions asked. Both the sample size and the depth of sequencing are sufficient, and the methods used to estimate evolutionary rates and the strength of selection are appropriate. The data presented are consistent with faster evolution of genes with male-biased expression, due to both positive and relaxed selection.
This is a useful contribution to understanding the effect of sex-biased expression in genetic evolution in plants. It demonstrates the range of variation in evolutionary rates and selective mechanisms, and provides further context to connect these patterns to potential explanatory factors in plant diversity such as the age of sex chromosomes and the developmental trajectories of male and female flowers.
Weaknesses:
The presence of sex chromosomes is a potential confounding factor, since there are different evolutionary expectations for X-linked, Y-linked, and autosomal genes. Attempting to distinguish transcripts on the sex chromosomes from autosomal transcripts could provide additional insight into the relative contributions of positive and relaxed selection.
Reviewer #3 (Public Review):
The potential for sexual selection and the extent of sexual dimorphism in gene expression have been studied in great detail in animals, but hardly examined in plants so far. In this context, the study by Zhao, Zhou et al. al represents a welcome addition to the literature.
Relative to the previous studies in Angiosperms, the dataset is interesting in that it focuses on reproductive rather than somatic tissues (which makes sense to investigate sexual selection), and includes more than a single developmental stage (buds + mature flowers).
Some aspects of the presentation have been improved in this new version of the manuscript.Specifically:
- the link between sex-biased and tissue-biased genes is now slightly clearer,
- the limitation related to the de novo assembled transcriptome is now formally acknowledged,
- the interpretation of functional categories of the genes identified is more precise,
- the legends of supplementary figures have been improved - a large number of typos have been fixed.
in response to this first round of reviews. As I detail below, many of the relevant and constructive suggestions by the previous reviewers were not taken into account in this revision.
For instance:
- Reviewer 2 made precise suggestions for trying to take into account the potential confounding factor of sex-chromosomes. This suggestion was not followed.
For the question of reviewer 2:
The presence of sex chromosomes is a potential confounding factor, since there are different evolutionary expectations for X-linked, Y-linked, and autosomal genes. Attempting to distinguish transcripts on the sex chromosomes from autosomal transcripts could provide additional insight into the relative contributions of positive and relaxed selection.
Empirically, the analyses could be expanded by an attempt to distinguish between genes on the autosomes and the sex chromosomes. Genotypic patterns can be used to provisionally assign transcripts to XY or XX-like behavior when all males are heterozygous and all females are homozygous (fixed X-Y SNPs) and when all females are heterozygous and males are homozygous (lost or silenced Y genes). Comparing such genes to autosomal genes with sex-biased expression would sharpen the results because there are different expectations for the efficacy of selection on sex chromosomes. See this paper (Hough et al. 2014; https://www.pnas.org/doi/abs/10.1073/pnas.1319227111), which should be cited and does in fact identify faster substitution rates in Y-linked genes.
Authors’ response: We have cited Hough et al. (2014) and Sandler et al. (2018) in the revised manuscript. We agree that the presence of sex chromosomes is potentially a confounding factor. By adopting methods in Hough et al. (2014) and Sandler et al. (2018), we tried to distinguish transcripts on sex chromosomes from autosomal chromosomes. For a total of 2,378 unbiased genes, we found that 36 genes were putatively sex chromosomal genes, 20 of which were exclusively heterozygous and homozygous for males and females, respectively; while the other 16 genes showing an opposite genotyping patterns between males and females. For 343 male-biased genes, only three ones exhibit a pattern of potentially sex-linked. For the 1,145 female-biased genes, we identified 19 genes which might located on the sex chromosomes. Among the 19 genes, five genes were exclusively heterozygous for males and exclusively homozygous for females, while reversed genotyping patterns presented in the other 14 genes. So, sex-linked genes may contribute relatively little to rapid evolution of male-biased genes. An alternative explanation is that the results could be unreliable due to small sample sizes. Thus, we did not describe them in the Results section. We will investigate the issue when whole genome sequences and population datasets become available in the near future.
- Reviewer 1 & 3 indicated that results were mentioned in the discussion section without having been described before. This was not fixed in this new version.
For the question of reviewer 1:
- Paragraph (407-416) describes the analysis of duplicated genes under relaxed selection but there is no mention of this in the results.
Authors’ response: Following this suggestion, in the Results section, we have added a sentence, “We also found that most of them were members of different gene families generated by gene duplication (Table S13)” on line 310-311 in the revised manuscript (Rapid_evolution_of_malebiased_genes_Trichosanthes_pilosa_Tracked_change_2023_11_06.docx).
For the question of reviewer 1:
38- line 417-424. The discussion should not contain new results.
Authors’ response: Thank you for pointing out this. In the Results section, we have added a few sentences as following: “Similarly, given that dN/dS values of sex-biased genes were higher due to codon usage bias, lower dS rates would be expected in sex-biased genes relative to unbiased genes (Ellegren & Parsch, 2007; Parvathy et al., 2022). However, in our results, the median of dS values in male-biased genes were much higher than those in female-biased and unbiased genes in the results of ‘free-ratio’ (Fig. S4A, female-biased versus male-biased genes, P = 6.444e-12 and malebiased versus unbiased genes, P = 4.564e-13) and ‘two-ratio’ branch model (Fig. S4B, femalebiased versus male-biased genes, P = 2.2e-16 and male-biased versus unbiased genes, P = 9.421e08, respectively). ” on line 323-331, and consequently, removed the following sentence, “femalebiased vs male-biased genes, P = 6.444e-12 and male-biased vs unbiased genes, P = 4.564e-13” and “female-biased versus male-biased genes, P = 2.2e-16 and male-biased versus unbiased genes, P = 9.421e-08, respectively” in the Discussion section.
- Reviewer 1 asked for a comparison between the number of de novo assembled unigenes in this transcriptome and the number of genes in other Cucurbitaceae species. I could not see this comparison reported.
Authors’ response: In the first revision, we described only percentages. We have now added the number of genes. We modify this part as follows: “The majority of unigenes were annotated by homologs in species of Cucurbitaceae (61.6%, 36,375), including Momordica charantia (16.3%, 9,625), Cucumis melo (11.9%, 7,027), Cucurbita pepo (11.9%, 7,027), Cucurbita moschata (11.5%, 6,791), Cucurbita maxima (10.1%, 5,964) and other species (38.4%, 22,676) (Fig. S1C).”.
- Reviewer 1 pointed out that permutation tests were more appropriate, but no change was made to the manuscript.
Authors’ response: Thank you for your suggestion. In the first revision, we have indirectly responded to the issues. Wilcoxon rank sum test is more commonly used for all comparisons between sex-biased and unbiased genes in many papers. Additionally, we tested datasets using permutation t-tests, which is consistent with the results of Wilcoxon rank sum test. For example, we found that only in floral buds, there are significant differences in ω values in the results of ‘free-ratio’ (female-biased versus male-biased genes, P = 0.04282 and male-biased versus unbiased genes, P = 0.01114) and ‘two-ratio’ model (female-biased versus male-biased genes, P = 0.01992 and male-biased versus unbiased genes, P = 0.02127, respectively). We also described these results in the Results section accordingly (line 278-284).
- Reviewer 3 pointed out the small sample size (both for the RNA-seq and the phylogenetic analysis), but again this limitation is not acknowledged very clearly.
Authors’ response: Sorry, we acknowledged that our sample size was relatively small. In the revised version, we have added a sentence as follows, “Additionally, our sample size is relatively small, and may provide low power to detect differential expression.” in the Discussion section.
- Reviewer 1 & 3 pointed out that Fig 3 was hard to understand and asked for clarifications that I did not see in the text and the figure in unchanged.
Authors’ response: Thank you for your suggestions. We have revised the manuscript to clarify the meaning of the acronym (F1TGs, F2TGs, M1TGs, M2TGs, F1BGs, F2BGs, M1BGs and M2BGs) and presented the number of genes. We have added two labels, indicating that panels A and B correspond to males and C and D to females in Fig. 3.
- Reviewer 3 suggested to combine all genes with sex-bias expression when evaluating the evolutionary rate, in addition to the analyses already done. This suggestion was not followed.
For the question of reviewer 3:line 196 and following: In these analyses, I could not understand the rationale for keeping buds vs mature flowers as separate analyses throughout. Why not combine both and use the full set of genes showing sex-bias in any tissue? This would increase the power and make the presentation of the results a lot more straightforward.
Authors’ response: Thank you for your suggestions. In the first revision, we tried to respond to the issues. First, we observed strong sexual dimorphism in floral buds, such as racemose versus solitary, early-flowering versus late-flowering. Second, as you pointed out earlier, “the dataset is interesting in that it focuses on reproductive rather than somatic tissues (which makes sense to investigate sexual selection), and includes more than a single developmental stage (buds + mature flowers)”, we totally agree with you on this point. Third, according to your suggestions, we combined all genes with sex-bias expression to evaluate the evolutionary rates. We found significant differences (please see a Figure below) in ω values in the results of ‘free-ratio’ (female-biased versus male-biased genes, P =0.005622 and male-biased versus unbiased genes, P = 0.001961) and ‘two-ratio’ model (female-biased versus male-biased genes, P = 0.008546 and male-biased versus unbiased genes, P = 0.009831, respectively) using Wilcoxon rank sum test. However, the significance is lower than previous results in floral buds due to sex-biased genes of mature flower joined, especially compared to the results of “free-ratio model”. Additionally, we also test all combined genes with sex-bias expression using permutation t-test. Unfortunately, there are no significant differences in ω values expect for male-biased versus unbiased genes in the results of ‘free-ratio’ model (P = 0.03034) and ‘two-ratio’ model (P = 0.0376), respectively. To a certain extent, the combination of all genes with sex-bias expression may cover the signals of rapid evolution of sex-biased genes in floral buds. Therefore, these results are not described in our manuscript. In the near future, we would like to make further investigations through more development stages of flowers and new technologies (e.g. Single-Cell method, See Murat et al., 2023) in each sex to consolidate the conclusion, and it is hoped that we could find more meaningful results.
Author response image 1.
- Reviewer 3 pointed out that hand-picking specific categories of genes was not statistically valid, and in fact not necessary in the present context. This was not changed.
For the question of reviewer3: removing genes on a post-hoc basis seems statistically suspicious to me. I don't think your analysis has enough power to hand-pick specific categories of genes, and it is not clear what this brings here. I suggest simply removing these analyses and paragraphs.
Authors’ response: Thank you for your suggestions. We have changed them accordingly. We removed a part of the following paragraph, “To confirm the contributions of positive selection and relaxed selection to rapid rates of male-biased genes in floral buds, we generated three datasets of OGs by excluding different sets of genes. Specifically, we excluded 18 relaxed selective male-biased genes (5.23%), 98 positively selected male-biased genes (28.57%), and 112 male-biased genes (32.65%) under positive and relaxed selection from 343 OGs (Fig. S4). We observed that after excluding male-biased genes under relaxed purifying selection, the median (0.264) decreased by 0.34% compared to the median (0.265) of all OGs (Fig. S4A-B). However, after excluding positively selected male-biased genes, the median (0.236) was reduced by 11% (Fig. S4A, C) in the results of ‘free-ratio’ branch model. This pattern was consistent with the results of ‘two-ratio’ branch model as well (Fig. S4E-G).” on line 290 to 300.
However, we kept the following paragraph, “We also analyzed female-biased and unbiased genes that underwent positive and relaxed selection in floral buds (Tables S6-S10). We identified 216 (18.86%) positively selected, and 69 (6.03%) relaxed selective female-biased genes from 1,145 OGs, respectively. Similarly, we found 436 (18.33%) positively selected, and 43 (1.81%) unbiased genes under relaxed selection from 2,378 OGs, respectively. Notably, male-biased genes have a higher proportion (10%) of positively selected genes compared to female-biased and unbiased genes. However, relaxed selective male-biased genes have a higher proportion (3.24%) than unbiased genes, but about 0.8% lower than that of female-biased genes.”. In this way, we can compare the proportion of sex-biased genes that have undergone positive selection and release selection among female-biased genes, unbiased genes and male-biased genes in floral buds in the Discussion section.
- Reviewer 1 asked for all data to be public, but I could not find in the manuscript where the link to the data on ResearchGate was provided.
Authors’ response: We have added a link in the Data Availability section.
- Reviewers 1 & 3 pointed out that since only two tissues were compared, the claims on pleiotropy should have been toned down, but no change was made to the text.
Authors’ response: Thank you for your suggestions. We revised “due to low pleiotropic constraints” to “due to low evolutionary constraints” and revised “low pleiotropy” to “low constraints”.
- Reviewer 1 asked for a clarification on which genes are plotted on the heatmap of Fig3C and an explanation of the color scale. No change was made.
Authors’ response: Sorry for the confusion. Actually, Reviewer 1 asked that “Fig. 2C, which genes are plotted on the heatmap and what is the color scale corresponding to?” In the previous revision, we have revised them (See Fig. 2 Sex-biased gene expression for floral buds and flowers at anthesis in males and females of Trichosanthes pilosa). Sex-biased genes (the union of sex-biased genes in F1, M1, F2 and M2) are plotted on the heatmap. The color gradient represents from high to low (from red to green) gene expression.
- Reviewer 1 asked for panel B in Fig S5 and S6 to be removed. They are still there. They asked for abbreviations to be explained in the legend of Fig S8. This was not done. They asked for details about columns headers. Such detailed were not added. They asked for more recent references on line 53-56: this was not done.
Authors’ response: We have removed panel B in Fig. S5 and S6. We explained abbreviations in text and Fig. S8. We added more details about the column headers in Supplementary Table S4, S5, S6, S7, S8, S9 and S10. We also added more recent references on line 53-56.
Recommendations for the authors:
Reviewer #3 (Recommendations For The Authors):
Authors’ response: Thank you for your suggestions. We have revised/fixed these issues following your concerns and suggestions.
Line 46-48 would be clearer as « Sexual dimorphism is the condition where sexes of the same species exhibit different morphological, ecological and physiological traits in gonochoristic animals and dioecious plants, despite male and female individuals sharing the same genome except for sex chromosomes or sex-determining loci »
Authors’ response: Thanks. We have revised it accordingly.
Line 50: replace «in both » by «between the two »
Authors’ response: We have revised it.
Line 51: « genes exclusively » -> « genes expressed exclusively »
Authors’ response: We have revised it.
Line 58: « in many animals » -> « in several animal species »
Authors’ response: We have revised it to “in some animal species”.
Line 58: « to which » -> « of this bias »
Authors’ response: We have revised it.
Line 64: « Most dioecious plants possess homomorphic sex-chromosomes that are roughly similar in size when viewed by light microscopy. » : a reference is missing
Authors’ response: We have added the reference.
Line 67: remove « that »
Authors’ response: We have revised it.
line 96: change to: « only the five above-mentioned studies »
Authors’ response: We have revised it.
Line 97: remove « the »
Authors’ response: We have revised it.
Line 111: « Drosophia » -> Drosophila
Authors’ response: We have revised it.
Line 114: exhibiting -> « exhibited »
Authors’ response: We have revised it.
Line 115: suggest -> « suggesting »
Authors’ response: We have revised it.
Line 117: « studies in plants have rarely reported elevated rates of sex-biased genes » : is it « rarely » or « never » ?
Authors’ response: We have revised to “never”.
Line 143: « It’s » -> « Its »
Authors’ response: We have revised it.
Line 143-146: say whether the male parts (e.g. anthers) are still present in females flowers, and the female parts (pistil+ ovaries) in the male flowers, or whether these respective organs are fully aborted.
Authors’ response: We have added the following sentence, “The male parts (e. g., anthers) of female flowers, and the female parts (e. g., pistil and ovaries) of male flowers are fully aborted” in line 148150 of the Introduction section.
Line 158: this is now clearer, but please specify whether you are talking about 12 floral buds in total, or 12 per individual (i.e. 72 buds in total).
Authors’ response: We have revised it to “Using whole transcriptome shotgun sequencing, we sequenced floral buds and flowers at anthesis from female and male of dioecious T. pilosa. We set up three biological replicates from three female and three male plants, including 12 samples in total (six floral buds and six flowers at anthesis)”.
Line 194-198: These sentences are unclear and hard to link to the figure. Consider changing for « In male plants, the number of tissue-biased genes in flowers at anthesis (M2TGs: n = 2795) was higher than that in floral buds (M1TGs: n = 1755, Fig. 3A and 3B). Figure 3 is also very hard to read. Adding a label on the side to indicate that panels A and B correspond to male-biased genes and C and D to female-biased genes could be useful.
Authors’ response: Thank you for your suggestions. We have revised the text to clarify the meaning of the acronym (F1TGs, F2TGs, M1TGs, M2TGs, F1BGs, F2BGs, M1BGs and M2BGs) and presented the number of genes. We have added two labels, indicating that panels A and B correspond to males and C and D to females in Figure 3.
Line 208: explain the approach: e.g. « We then compared rates of protein evolution among malebiased, female-biased and unbiased genes. To do this, we sequenced floral bud transcriptomes from the closely related T. anguina, as well as two more distant outgroups, T. kirilowii and Luffa cylindrica. T. kirilowii is a dioecious species like T. pilosa, and the other two are monoecious. We identified one-to-one orthologous groups (OGs) for 1,145 female-biased, 343 male-biased, and 2,378 unbiased genes. »
Authors’ response: We have revised this paragraph to the following, “We compared rates of protein evolution among male-biased, female-biased and unbiased genes in four species with phylogenetic relationships (((T. anguina, T. pilosa), T. kirilowii), Luffa cylindrica), including dioecious T. pilosa, dioecious T. kirilowii, monoecious T. anguina in Trichosanthes, together with monoecious Luffa cylindrica. To do this, we sequenced transcriptomes of T. pilosa. We also collected transcriptomes of T. kirilowii, as well as genomes of T. anguina and Luffa cylindrica.”
Line 220: « the same ω value was in all branches » -> « all branches are constrained to have the same ω value ».
Authors’ response: We have revised it.
Line 221: « results of the 'two-ratio' branch model ... »
Authors’ response: We have revised it.
Line 235: add a few words to explain why the effect size is bigger than for buds, but still is not significant: e.g. «possibly because of limited statistical power due to the low number of sex-biased genes in flowers at anthesis »
Authors’ response: We have revised this to “However, there is no statistically significant difference in the distribution of ω values using Wilcoxon rank sum tests for female-biased versus male-biased genes (P = 0.0556), female-biased versus unbiased genes (P = 0.0796), and male-biased versus unbiased genes (P = 0.3296) possibly because of limited statistical power due to the low number of sex-biased genes in flowers at anthesis.” in line 260-261.
Line 255: explain in plain English what the « A model » is. This was already requested in the previous version.
Authors’ response: We have revised “A model” to “classical branch-site model A”.
Line 258: explain in plain English what the « foreground 2b ω value » corresponds to
Authors’ response: We have revised to as follows, “foreground 2b ω value” to “foreground ω >1”. Additionally, we also added the sentence “The classical branch-site model assumes four site classes (0, 1, 2a, 2b), with different ω values for the foreground and background branches. In site classes 2a and 2b, the foreground branch undergoes positive selection when there is ω > 1.” in line 624-627.
Line 259: explain how these different approaches complement each other rather than being redundant. This was also already requested in the previous version.
Authors’ response: Sorry. We have now revised it as follows, “As a complementary approach, we utilized the aBSREL and BUSTED methods that are implemented in HyPhy v.2.5 software, which avoids false positive results by classical branch-site models due to the presence of rate variation in background branches, and detected significant evidence of positive selection.” in line 292-295.
Line 270: remove « dramatically », and also remove « or eliminated at both gene-wide and genomewide levels », as well as « relative to positive selection »
Authors’ response: Thank you for your suggestions. We have revised it.
Line 290-309: remove this section - this was already pointed out in the previous reviews as a « ad hoc » procedure, and this point has already been made clear with the RELAX analysis.
Authors’ response: Thank you for your suggestions. We revised this section accordingly. We remove the following paragraph, “To confirm the contributions of positive selection and relaxed selection to rapid rates of male-biased genes in floral buds, we generated three datasets of OGs by excluding different sets of genes. Specifically, we excluded 18 relaxed selective male-biased genes (5.23%), 98 positively selected male-biased genes (28.57%), and 112 male-biased genes (32.65%) under positive and relaxed selection from 343 OGs (Fig. S4). We observed that after excluding malebiased genes under relaxed purifying selection, the median (0.264) decreased by 0.34% compared to the median (0.265) of all OGs (Fig. S4A-B). However, after excluding positively selected malebiased genes, the median (0.236) was reduced by 11% (Fig. S4A, C) in the results of ‘free-ratio’ branch model. This pattern was consistent with the results of ‘two-ratio’ branch model as well (Fig. S4E-G).” on line 334-344.
However, we kept the other parts “We also analyzed female-biased and unbiased genes that underwent positive and relaxed selection in floral buds (Tables S6-S10). We identified 216 (18.86%) positively selected, and 69 (6.03%) relaxed selective female-biased genes from 1,145 OGs, respectively. Similarly, we found 436 (18.33%) positively selected, and 43 (1.81%) unbiased genes under relaxed selection from 2,378 OGs, respectively. Notably, male-biased genes have a higher proportion (10%) of positively selected genes compared to female-biased and unbiased genes. However, relaxed selective male-biased genes have a higher proportion (3.24%) than unbiased genes, but about 0.8% lower than that of female-biased genes.”. In this way, we can compare the proportion of sex-biased genes that have undergone positive selection and release selection among female-biased genes, unbiased genes and male-biased genes in floral buds in the Discussion sections.
Line 348: Here you talk about « Numerous studies », but then only report three studies. Please clarify.
Authors’ response: Thank you for your suggestions. We have revised it to “Several studies”.
Line 352: Cut the sentence: « In contrast, the wind-pollinated dioecious plant Populus balsamifera ... »
Authors’ response: Thank you for your suggestions. We have revised it.
Line 357: « In contrast to the above studies... »: If I understand correctly, this is not in contrast to the observation in Populus balsamifera. Please clarify.
Authors’ response: Thank you for your suggestions. We have revised to “Similar to the above study of Populus balsamifera.”.
Line 420: « our results » -> « we »; « that underwent » -> « undergoing »
Authors’ response: Thank you for your suggestions. We have revised it.
Figure 3 is very hard to read and poorly labeled (see my comments on line 194 above). It is also hard to link to the text, since the numbers reported in the text are actually not present in the figure unless the readers makes some calculations themselves. This should be improved. Also, the use of acronyms (e.g. M1BG, F2TG etc.) contributes to making the text very difficult to read. The acronyms should at least be explained very clearly in the text when they are used.
Authors’ response: Thank you for your suggestions. We have revised the text to clarify the meaning of the acronym (F1TGs, F2TGs, M1TGs, M2TGs, F1BGs, F2BGs, M1BGs and M2BGs) and give the number of genes. We have added two labels, indicating that panels A and B correspond to males and C and D to females in Figure 3.
-

-

eLife assessment
This valuable paper examines gene expression differences between male and female individuals over the course of flower development in the dioecious angiosperm Trichosantes pilosa. Male-biased genes evolve faster than female-biased and unbiased genes, which is frequently observed in animals, but this is the first report of such a pattern in plants. In spite of the limited sample size, the evidence is mostly solid and the methods appropriate for a non-model organism. The resources produced will be used by researchers working in the Cucurbitaceae, and the results obtained advance our understanding of the mechanisms of plant sexual reproduction and its evolutionary implications: as such they will broadly appeal to evolutionary biologists and plant biologists.
-

Reviewer #1 (Public Review):
The evolution of dioecy in angiosperms has significant implications for plant reproductive efficiency, adaptation, evolutionary potential, and resilience to environmental changes. Dioecy allows for the specialization and division of labor between male and female plants, where each sex can focus on specific aspects of reproduction and allocate resources accordingly. This division of labor creates an opportunity for sexual selection to act and can drive the evolution of sexual dimorphism.
In the present study, the authors investigate sex-biased gene expression patterns in juvenile and mature dioecious flowers to gain insights into the molecular basis of sexual dimorphism. They find that a large proportion of the plant transcriptome is differentially regulated between males and females with the number of …
Reviewer #1 (Public Review):
The evolution of dioecy in angiosperms has significant implications for plant reproductive efficiency, adaptation, evolutionary potential, and resilience to environmental changes. Dioecy allows for the specialization and division of labor between male and female plants, where each sex can focus on specific aspects of reproduction and allocate resources accordingly. This division of labor creates an opportunity for sexual selection to act and can drive the evolution of sexual dimorphism.
In the present study, the authors investigate sex-biased gene expression patterns in juvenile and mature dioecious flowers to gain insights into the molecular basis of sexual dimorphism. They find that a large proportion of the plant transcriptome is differentially regulated between males and females with the number of sex-biased genes in floral buds being approximately 15 times higher than in mature flowers. The functional analysis of sex-biased genes reveals that chemical defense pathways against herbivores are up-regulated in the female buds along with genes involved in the acquisition of resources such as carbon for fruit and seed production, whereas male buds are enriched in genes related to signaling, inflorescence development and senescence of male flowers. Furthermore, the authors implement sophisticated maximum likelihood methods to understand the forces driving the evolution of sex-biased genes. They highlight the influence of positive and relaxed purifying selection on the evolution of male-biased genes, which show significantly higher rates of non-synonymous to synonymous substitutions than female or unbiased genes. This is the first report (to my knowledge) highlighting the occurrence of this pattern in plants. Overall, this study provides important insights into the genetic basis of sexual dimorphism and the evolution of reproductive genes in Cucurbitaceae.
-

Reviewer #2 (Public Review):
Summary:
This study uses transcriptome sequence from a dioecious plant to compare evolutionary rates between genes with male- and female-biased expression and distinguish between relaxed selection and positive selection as causes for more rapid evolution. These questions have been explored in animals and algae, but few studies have investigated this in dioecious angiosperms, and none have so far identified faster rates of evolution in male-biased genes (though see Hough et al. 2014 https://doi.org/10.1073/pnas.1319227111).
Strengths:
The methods are appropriate to the questions asked. Both the sample size and the depth of sequencing are sufficient, and the methods used to estimate evolutionary rates and the strength of selection are appropriate. The data presented are consistent with faster evolution of …
Reviewer #2 (Public Review):
Summary:
This study uses transcriptome sequence from a dioecious plant to compare evolutionary rates between genes with male- and female-biased expression and distinguish between relaxed selection and positive selection as causes for more rapid evolution. These questions have been explored in animals and algae, but few studies have investigated this in dioecious angiosperms, and none have so far identified faster rates of evolution in male-biased genes (though see Hough et al. 2014 https://doi.org/10.1073/pnas.1319227111).
Strengths:
The methods are appropriate to the questions asked. Both the sample size and the depth of sequencing are sufficient, and the methods used to estimate evolutionary rates and the strength of selection are appropriate. The data presented are consistent with faster evolution of genes with male-biased expression, due to both positive and relaxed selection.
This is a useful contribution to understanding the effect of sex-biased expression in genetic evolution in plants. It demonstrates the range of variation in evolutionary rates and selective mechanisms, and provides further context to connect these patterns to potential explanatory factors in plant diversity such as the age of sex chromosomes and the developmental trajectories of male and female flowers.
Weaknesses:
The presence of sex chromosomes is a potential confounding factor, since there are different evolutionary expectations for X-linked, Y-linked, and autosomal genes. Attempting to distinguish transcripts on the sex chromosomes from autosomal transcripts could provide additional insight into the relative contributions of positive and relaxed selection.
-

Reviewer #3 (Public Review):
The potential for sexual selection and the extent of sexual dimorphism in gene expression have been studied in great detail in animals, but hardly examined in plants so far. In this context, the study by Zhao, Zhou et al. al represents a welcome addition to the literature.
Relative to the previous studies in Angiosperms, the dataset is interesting in that it focuses on reproductive rather than somatic tissues (which makes sense to investigate sexual selection), and includes more than a single developmental stage (buds + mature flowers).
-

-

Author Response
The following is the authors’ response to the original reviews.
Thank you for reviewing our manuscript. We do find that the reviews are constructive and meaningful. Accordingly, we incorporated most suggestions into our revision. We provided a point-by-point responses to the reviews below.
Reviewer #1 (Public Review):
The evolution of dioecy in angiosperms has significant implications for plant reproductive efficiency, adaptation, evolutionary potential, and resilience to environmental changes. Dioecy allows for the specialization and division of labor between male and female plants, where each sex can focus on specific aspects of reproduction and allocate resources accordingly. This division of labor creates an opportunity for sexual selection to act and can drive the evolution of sexual dimorphism.
In the present …
Author Response
The following is the authors’ response to the original reviews.
Thank you for reviewing our manuscript. We do find that the reviews are constructive and meaningful. Accordingly, we incorporated most suggestions into our revision. We provided a point-by-point responses to the reviews below.
Reviewer #1 (Public Review):
The evolution of dioecy in angiosperms has significant implications for plant reproductive efficiency, adaptation, evolutionary potential, and resilience to environmental changes. Dioecy allows for the specialization and division of labor between male and female plants, where each sex can focus on specific aspects of reproduction and allocate resources accordingly. This division of labor creates an opportunity for sexual selection to act and can drive the evolution of sexual dimorphism.
In the present study, the authors investigate sex-biased gene expression patterns in juvenile and mature dioecious flowers to gain insights into the molecular basis of sexual dimorphism. They find that a large proportion of the plant transcriptome is differentially regulated between males and females with the number of sex-biased genes in floral buds being approximately 15 times higher than in mature flowers. The functional analysis of sex-biased genes reveals that chemical defense pathways against herbivores are up-regulated in the female buds along with genes involved in the acquisition of resources such as carbon for fruit and seed production, whereas male buds are enriched in genes related to signaling, inflorescence development and senescence of male flowers. Furthermore, the authors implement sophisticated maximum likelihood methods to understand the forces driving the evolution of sexbiased genes. They highlight the influence of positive and relaxed purifying selection on the evolution of male-biased genes, which show significantly higher rates of nonsynonymous to synonymous substitutions than female or unbiased genes. This is the first report (to my knowledge) highlighting the occurrence of this pattern in plants. Overall, this study provides important insights into the genetic basis of sexual dimorphism and the evolution of reproductive genes in Cucurbitaceae.
Thank you for your positive comments. Greatly appreciated.
There are, however, parts of the manuscript that are not clearly described or could be otherwise improved.
- The number of denovo-assembled unigenes seems large and I would like to know how it compares to the number of genes in other Cucurbitaceae species. The presence of alternatively assembled isoforms or assembly artifacts may be still high in the final assembly and inflate the numbers of identified sex-biased genes.
The majority of unigenes were annotated by homologs in species of Cucurbitaceae (63%), including Momordica charantia (16.3%), Cucumis melo (11.9%), Cucurbita pepo (11.9%), Cucurbita moschata (11.5%), Cucurbita maxima (10.1%) and other species of Cucurbitaceae (Fig. S1C). We admit that in the final assembly, transcripts may be still overestimated due to the unavoidable presence of isoforms, although we have tried our best to filter it by several strategies of clustering methods. Additionally, we assessed the transcripts using BUSCOv5.4.5 and embryophyta_odb10 database with 1,614 plant orthologs assessment. Some 95.0% of these orthologs were covered by the unigenes, in which 1447 (89.7%) BUSCO genes were “Complete BUSCOs”, 85 (5.3%) were “Fragmented BUSCOs”, and only 82 (5.0%) were “Missing BUSCOs” (Table S2). Overall, our assessment suggested that we have generated high-quality reference transcriptomes in the absence of a reference genome. Subsequently, we revised the manuscript (lines 175-181).
- It is interesting that the majority of sex-biased genes are present in the floral buds but not in the mature flowers. I think this pattern could be explored in more detail, by investigating the expression of male and female sex-biased genes throughout the flower development in the opposite sex. It is also not clear how the expression of the sex-biased genes found in the buds changes when buds and mature flowers are compared within each sex.
Thank you for your advice for further understanding of this interesting pattern. In the near future, we would like to study these issues through more development stages of flowers in each sex, probably with the aid of single-cell techniques and a reference genome. We have revised the manuscript to reflect these in Results, in the section "Tissue-biased/stage-biased gene expression" (lines 202216).
- The statistical analysis of evolutionary rates between male-biased, female-biased, and unbiased genes is performed on samples with very different numbers of observations, therefore, a permutation test seems more appropriate here.
Thank you for your suggestion. However, all comparisons between sex-biased and unbiased genes were tested using Wilcoxon rank sum test in R software, which is more commonly used. Additionally, we tested some datasets, which were consistent with Wilcoxon rank sum test.
- The impact of pleiotropy on the evolutionary rates of male-biased genes is speculative since only two tissue samples (buds and mature flowers) are used. More tissue types need to be included to draw any meaningful conclusions here.
Thank you for your advice for further understanding of the impact of pleitropy. In the near future, we would like make further investigations through more development stages of flowers and new technologies in each sex to consolidate the conclusion.
Reviewer #2 (Public Review):
Summary:
This study uses transcriptome sequence from a dioecious plant to compare evolutionary rates between genes with male- and female-biased expression and distinguish between relaxed selection and positive selection as causes for more rapid evolution. These questions have been explored in animals and algae, but few studies have investigated this in dioecious angiosperms, and none have so far identified faster rates of evolution in male-biased genes (though see Hough et al. 2014 https://doi.org/10.1073/pnas.1319227111).
Strengths:
The methods are appropriate to the questions asked. Both the sample size and the depth of sequencing are sufficient, and the methods used to estimate evolutionary rates and the strength of selection are appropriate. The data presented are consistent with faster evolution of genes with male-biased expression, due to both positive and relaxed selection.
This is a useful contribution to understanding the effect of sex-biased expression in genetic evolution in plants. It demonstrates the range of variation in evolutionary rates and selective mechanisms, and provides further context to connect these patterns to potential explanatory factors in plant diversity such as the age of sex chromosomes and the developmental trajectories of male and female flowers.
Weaknesses:
The presence of sex chromosomes is a potential confounding factor, since there are different evolutionary expectations for X-linked, Y-linked, and autosomal genes. Attempting to distinguish transcripts on the sex chromosomes from autosomal transcripts could provide additional insight into the relative contributions of positive and relaxed selection.
Thank you for your meanful suggestions. We agree that the identification of chromosome origins for transcripts would greatly improve the insights of selection, and we will investigate these issues, probably with a reference genome in the near future.
Reviewer #3 (Public Review):
The potential for sexual selection and the extent of sexual dimorphism in gene expression have been studied in great detail in animals, but hardly examined in plants so far. In this context, the study by Zhao, Zhou et al. al represents a welcome addition to the literature.
Relative to the previous studies in Angiosperms, the dataset is interesting in that it focuses on reproductive rather than somatic tissues (which makes sense to investigate sexual selection), and includes more than a single developmental stage (buds + mature flowers).
The main limitation of the study is the very low number of samples analyzed, with only three replicate individuals per sex (i.e. the whole study is built on six individuals only). This provides low power to detect differential expression. Along the same line, only three species were used to evaluate the rates of non-synonymous to synonymous substitutions, which also represents a very limited dataset, in particular when trying to fit parameter-rich models such as those implemented here.
A third limitation relates to the absence of a reference genome for the species, making the use of a de novo transcriptome assembly necessary, which is likely to lead to a large number of incorrectly assembled transcripts. Of course, the production of a reference transcriptome in this non-model species is already a useful resource, but this point should at least be acknowledged somewhere in the manuscript.
Each of these shortcomings is relatively important, and together they strongly limit the scope of the conclusions that can be made, and they should at least be acknowledged more prominently. The study is valuable in spite of these limitations and the topic remains grossly understudied, so I think the study will be of interest to researchers in the field, and hopefully inspire further, more comprehensive analyses.
We acknowledged that our sample size was relatively small. We will investigate these issues at the population level, probably with a reference genome in the near future. We acknowledged in the revised manuscript that there may be some incorrectly assembled transcripts. We assessed the transcripts using BUSCOv5.4.5 and the latest embryophyta_odb10 database with 1,614 plant orthologs assessment. As mentioned, 95.0% of these orthologs were covered by the unigenes, which of 1447 (89.7%) BUSCO genes were “Complete BUSCOs”, 85 (5.3%) were “Fragmented BUSCOs”, and only 82 (5.0%) were “Missing BUSCOs” (Table S2). In short, the quality of transcriptome was high in the absence of a reference genome.
Reviewer #1 (Recommendations For The Authors):
My main criticism of this manuscript is that it refers to gene names and orthogroups throughout the text, however, the assembled transcripts are not accessible. The reference trascriptome, orthology data, and alignments used for evolutionary analysis should be made available through a public repository to support reproducibility and efficient use of produced resources in this study.
We have uploaded these datasets in Researchgate (https://www.researchgate.net/publication/373194650_Trichosanthes_pilosa_datasets Positive_selection_and_relaxed_purifying_selection_contribute_to_rapid_evolution of_male-biased_genes_in_a_dioecious_flowering_plant).
Comments to the authors:
- I have an issue with the tissue-biased gene expression analysis. Looking at Fig.3, it seems to me there are 3,204 male-biased genes that are expressed at the same level in male buds and mature flowers (same for 5,011 female-biased genes in female buds and flowers), however, only a handful of genes show sex bias between mature male and female flowers. Taking the male-biased genes as an example, if the 3,204 M1BGs experience the same expression levels in mature male flowers and are no longer male-biased when mature male vs female flowers are compared, why there are not found as female tissue biased (F2TGs)? I may be wrong, but one scenario would be that the M1BGs increase their expression in female flowers and become unbiased. However, that increase in expression (low expression in the female buds → higher expression in the female flowers) should classify them as female tissue-biased genes (F2TGs). Can you please clarify how are the M1BGs and F1BGs expressed in the flowers of the opposite sex?
As to Fig. 3A, 3,204 male-biased genes expressed in male floral buds are part of all male-biased genes (3204+286+724=4214), as shown in Fig.2A. However, only 233 male-biased genes (88+1+144=233, Fig.2B and Fig.3B) expressed in male mature flowers. So, they are not expressed at the same level between male floral buds and mature flowers. Only 288 genes are sex-biased (M1BGs), as well as tissue/stage-biased (M1TGs) in male floral buds. M1BGs (4,214 male-biased genes) and F1BGs (5,096 female-biased genes) are 0 overlaps, except for 44,326 unbiasedgenes shown in Fig.2A. That is, F1BGs (5,096 female-biased genes) are low expression or no expression in M1BGs (4,214 male-biased genes). The expression levels of some genes have been shown in Table S14.
- Paragraph (407-416) describes the analysis of duplicated genes under relaxed selection but there is no mention of this in the results.
In fact, these results have been shown in Table S13. It is not necessary for us to describe them in detail in the results.
- How did the authors conclude that the identified functions in male flowers make them more adapted to biotic and abiotic environments (line 347-350)? In the paragraph above (line 338-342) the authors describe that female buds are better equipped against herbivores, which are a biotic factor?
Following your concerns, we have revised the manuscript as follows: For line 338-342, we revised the text as “Indeed, functional enrichment analysis in chemical pathways such as terpenoid backbone and diterpenoid biosynthesis indicated that relative to male floral buds, female floral buds had more expressed genes that were equipped to defend against herbivorous insects and pathogens, except for growth and development (Vaughan et al., 2013; Ren et al., 2022) (Fig. S7A and Table S11).” For line 347-350, we revised text as “We also found that male-biased genes with high evolutionary rates in male buds were associated with functions to abiotic stresses and immune responses (Tables S12 and S13), which suggest that male floral buds through rapidly evolving genes are adapted to mountain climate and the environment in Southwest China compared to female floral buds through high gene expression.”
- Line 417-418: decreasing codon usage bias is linked to decreasing synonymous substitution rates, should this be the opposite?
No. Codon usage bias was positively related to synonymous substitution rates. That is, stronger codon usage bias may be related to higher synonymous substitution rates (Parvathy et al., 2022).
- Figures and Tables are not standalone and are missing details in the legends. - Fig.2C, which genes are plotted on the heatmap and what is the color scale corresponding to?
- All Supplementary figures are missing the descriptions of individual panels (A, B, C,etc.) in the legends. In addition, please add the numbers of observations under boxplots.
- Supplementary Fig.5 and 6: Panel B is not a Venn diagram, I suggest removing it from the figures.
- Supplementary Fig.7: Should be 'sex-biased genes'. What is the x-axis on the plot?
- Supplementary Fig.8: Please add the description of the abbreviations in the legend. - Supplementary Tables S4, S5, S6: Please add information about the foreground and background branches.
- Supplementary Table S6, S7, S8, S9, S10: Please add more details about the column headers (what is Model-A, background ω 2a, Unconstrained_1.p, K, which was the foreground branch etc.).
- Supplementary Table S11: Please add gene IDs for each KEGG category.
We have revised/fixed these issues following your concerns and suggetions.
Minor comments:
Line 28: 'algae' in place of 'algas'
Line 53-56: Please provide more recent references.
Line65: 'most' instead of 'almost'
Line 86-87: It is not clear from the sentence if the sex-biased expression was detected in flowers compared to leaves, or were the sex-biased genes detected between male and female leaves? Please clarify.
Line 107-108: positive selection is referred to as adaptive evolution, please choose one or the other.
Line 109: 'force' instead of 'forces'
Line 110: 'algae' instead of 'alga'
Line 132: '..mainly distributed from Southwest,' the country is missing.
Line 202: 'protein sequence evolution'?
Line 232: what does the 'number of evolutionary rates' refers to?
Line 253: please provide a reference for the RELAX model.
Line 274: 'relaxed selective male-biased genes' should be 'male-biased genes under relaxed purifying selection'?
Line 318: Please add a sentence explaining why the Cucurbitaceae family is a great model to study the evolution of sexual systems.
Line 321: 'genes' instead of 'gene'.
Line 366: male-biased genes experience 'higher' or 'more rapid' evolutionary rates. line 377: in the present study and in the case of Ectocarpus alga, positive selection plays an important role in male-biased genes evolution, but does not account for the majority of evolutionary change. Therefore, I would not call it a 'primary' force.
Line 477: missing reference for DESeq2 package.
Line 480: 'used'.
Line 498: 'coding sequences'.
Line516: 'to' instead of 'by'.
Line 553: 'the' is repeated twice.
Sorry for the typos and grammatical issues. We have revised them accordingly.
Reviewer #2 (Recommendations For The Authors):
There are two areas for improvement, one empirical and one theoretical.
Empirically, the analyses could be expanded by an attempt to distinguish between genes on the autosomes and the sex chromosomes. Genotypic patterns can be used to provisionally assign transcripts to XY or XX-like behavior when all males are heterozygous and all females are homozygous (fixed X-Y SNPs) and when all females are heterozygous and males are homozygous (lost or silenced Y genes). Comparing such genes to autosomal genes with sex-biased expression would sharpen the results because there are different expectations for the efficacy of selection on sex chromosomes. See this paper (Hough et al. 2014; https://www.pnas.org/doi/abs/10.1073/pnas.1319227111), which should be cited and does in fact identify faster substitution rates in Y-linked genes (and note that pollenexpressed genes, at least, are concentrated on the sex chromosome in this system: https://academic.oup.com/evlett/article/2/4/368/6697528, https://royalsocietypublishing.org/doi/10.1098/rstb.2021.0226).
We have cited Hough et al. 2014 and noticed that several species have been observed to exhibit rapid evolutionary rates of sequences on sex chromosomes compared to autosomes, which has been related to the evolutionary theories of fast-X or fast-Z (lines 482-484).
On the theoretical side, this study is making a very specific intervention, namely identifying more rapid evolutionary rates in genes with male-biased than femalebiased expression in a dioecious plant. The writing in the introduction and the discussion needs to be improved to differentiate between this comparison and similar comparisons, e.g. sex-biased expression in other dioecious plants (76-81), between Xlinked and Y-linked genes (Hough et al. 2014), sex chromosomes and autosome (several studies already cited), gametophytic and sporophytic tissue, and male and female reproductive tissue in hermaphroditic plants. Setting out this distinction early in the introduction will make the specific goals and novelty of this work clearer.
Thank you for your constructive suggestions. We have revised the relevant part of the Introduction accordingly (lines 74-107).
Specific comments by line:
Sorry for the typos or wording issues. We have revised them.
26 - driven not driving
28 - check house style (algae vs algas)
28-29 - consider clarifying the antecedent of "them" (evolutionary forces, not algas) 35 - maybe, but don't the signalling genes involved in stress responses function in many capacities, not just stress? Also, there's evidence that reproductive recognition machinery in plants may ultimately derive from immune function (e.g. https://doi.org/10.1111/j.1469-8137.2008.02403.x), so the GO category "biotic stress" may be too vague
39 - maybe clarify that "for the first time" refers to male rather than female, since there have been other studies in dioecious plants
66-68 - asserting that something is "essential" after describing how rare it is doesn't quite follow, since diecious plants - especially with sex chromosomes - are basically an exception. I agree that understanding the evolution of dioecious plants is important, but this isn't the most compelling way to make that case - perhaps try something else.
137ff - this sentence can be consolidated and streamlined
142 - "floral tissue" rather than "flowers tissue," here and elsewhere
144 - divergence (singular)
235 - "evidence for the contributions of" = "evidences" is unidiomatic 250 - efficiency or efficacy?
300 - why is "inositol" capitalized here and elsewhere?
300ff - are these typical patterns in male tissue in other species?
308 - is that interesting? It seems like exactly what I'd expect. Perhaps start with the unsurprising but reassuring observation (anther and pollen development genes are indeed expressed in male buds) before moving on to the more surprising findings.
319 - remove "the"
321 - genes (plural)
330 - replace "these differences" with "the differences" 336 - perhaps recap proportions / percents here?
340 - unnecessary comma after diterpenoid
341 - this seems like a big leap from the evidence, especially in the absence of supporting information about the chemical defenses of these species and how they differ by sex. Don't terpenoids have a diverse array of functions, not just defense? Here's a review: https://link.springer.com/chapter/10.1007/10_2014_295
We have revised the text as “Indeed, functional enrichment analysis in chemical pathways such as terpenoid backbone and diterpenoid biosynthesis indicated that relative to male floral buds, female floral buds had more expressed genes that were equipped to defend against herbivorous insects and pathogens, except for growth and development (Vaughan et al., 2013; Ren et al., 2022) (Fig. S7A and Table S11)” (lines 373-378).
349 - as mentioned in line 35, this is a big speculative leap. The discussion is the place for speculation, but consider other explanations too. How does the development of flowers work? Are male flowers suppressing or resorbing female primordial organs? Do male flowers in fact senesce faster? perhaps spell out the logic in more detail.
We have revised the text as “In addition, the enrichment in regulation of autophagy pathways could be associated with gamete development and the senescence of male floral buds (Table S14) (Liu and Bassham, 2012; Li et al., 2020; Zhou et al., 2021). In fact, it was observed that male flowers senesced faster (Wu et al., 2011). We also found that homologous genes of two male-biased genes in floral buds (Table S14) that control the raceme inflorescence development (Teo et al., 2014) were highly expressed compared to female floral buds. Taken together, these results indicate that expression changes in sex-biased genes, rather than sex-specific genes play different roles in sexual dimorphic traits in physiology and morphology (Dawson and Geber, 1999).” (lines 390-402).
351 - senescence of, not senescence for
363 - but Hough et al. 2014 did show rapid evolution of Y-linked genes, and those are by definition sex biased ...
391 - perhaps reiterate here that while some sex-BIASED genes did, sex-SPECIFIC genes did not, to avoid confusion
We also revised them accordingly.
Reviewer #3 (Recommendations For The Authors):
1- lines 56-57 : « have facilitated » : this wording confounds correlation with causation. Consider rephrasing as « is associated with »
2- lines 58-60 : vague wording : what are these variations ? e.g. which tissues and stages are generally enriched?
3- line 63 : this sentence is a bit misleading: consider changing it to « Most dioecious plants possess homomorphic sex-chromosomes » [and explain what homomorphic means in this context].
4- line 68 : a reference is missing here. Also perhaps, allude to the fact that sexual selection in plants has long been considered a contentious issue (e.g. https://doi.org/10.1016/j.cub.2010.12.035)
5- lines 72-76 : beyond simply describing the pattern, say what evolutionary processes are revealed by these observations.
6- line 92 : remind the reader what these 5 studies are.
7- line 94-95 : explain why the comparison of vegetative vs vegetative and vegetative vs reproductive tissues is a problem.
The published studies only compared gene expression in vegetative versus vegetative tissues and vegetative versus reproductive tissues. Because it limited our understanding of sexual selection at different floral development stages. Revised accordingly (lines 103-104). We are very interested in flower development stage for sex-biased genes. The datasets could investigate sexual selection using two developmental stage (buds + mature flowers).
8- line 100 « Evolutionary dynamic analyses » : this wording is vague
9- line 110 : brown algae are NOT plants
10- line 137-140 or in M&M : needs to describe somewhere how the male flowers differ from the female flowers and vice-versa: are the whole morphological structures related to female (male) reproduction entirely missing, or is their development arrested later on and they are still present but simply not producing gametes? This has consequences for the interpretation of the genes they express.
We have revised the typos or wording issues accordingly. However, because the sampled floral buds were equal or less than 3 mm in size, we did not observe much morphological structural difference. Indeed, the male and female flowers at antheses were markedly different in this dioecious plant as shown in Fig. 1. Additionally, we found that dioecy is the ancestral state of Trichosanthes, and transitions to monoecy (Guo et al., 2020) based on our analysis (not shown in this study), which suggest that in the early stages of flower development, female floral buds may tend to masculinize, and vice versa (Fig. 2C).
11- line 152 : it is important to be very transparent on the sample sizes here: « from three females and three males of the dioecious ... »
12- line 153 : along the same line, explain here why a de novo transcriptome had to be generated here: « In the absence of an assembled reference genome for this nonmodel species, we de novo assembled ... »
13- line 164-165 : « we have generated high-quality reference trancriptomes » : I am not entirely convinced of the quality of the transcriptome obtained without a reference genome, so I suggest simply removing this subjective sentence.
Our assessment suggested that we have generated high-quality reference transcriptomes in the absence of a reference genome, which will be the next step of our work.
14- line 169 : briefly explain the criteria used to call differentially expressed genes. Given the threshold (log-fold change >=1.3 if I read the figure correctly, but the M&M says >=1), explain how it was chosen.
Sorry, you may have misunderstood the X, Y coordinates. The value of y coordinate represents -log10(FDR), and the value of x coordinate represents log2 (Fold Change).
15- line 174 : Not clear to me how Fig2C is « revealing strong sexual dimorphism », since genes cluster neither by sex nor by tissue. This should be explained more clearly.
16- line 174-177 : The fact that more ex-biased genes were identified in early buds than in mature flowers is an interesting observation that could be given more prominence in the manuscript, but it is not really explained. Could it reflect the fact that more genes are expressed in early buds because meiotic processes happen early in flower development? Also, the genes involved in male and female organ cell fate determination might also be expected to be expressed early, with mostly organ growth genes being expressed in the mature flower.
17- line 181 : a wrap-up sentence might be useful here to drive the point home that sex-bias is more prevalent in buds than mature flowers.
18- line 184 : « tissue-biased » : a more appropriate wording here would be « stagebiased », no ? These are indeed the same tissues but at different developmental stages.
19- line 183-195 : this section could benefit from setting clear expectations in a hypothesis testing framework laying out the reasons to expect a different bias between stages and between sexes. How does that fit with the level of morphological divergence between sexes (relates to my point 10 above).
20- line 197-204. A number of essential pieces of information are missing here: how many species, how divergent, say that one other is dioecious, and precise their relative phylogenetic placement (which is important to understand the models used below). Maybe adding a phylogeny of these species in Figure 4 could be useful. Also, briefly explain the « two-ratio » and « free-ratio » models here.
21- line 196 and following: In these analyses, I could not understand the rationale for keeping buds vs mature flowers as separate analyses throughout. Why not combine both and use the full set of genes showing sex-bias in any tissue? This would increase the power and make the presentation of the results a lot more straightforward.
As you pointed earlier (in the public review, paragraphy 2), “the dataset is interesting in that it focuses on reproductive rather than somatic tissues (which makes sense to investigate sexual selection), and includes more than a single developmental stage (buds + mature flowers)”, we totally agree with your points and were very interested in floral development stages for sex-biased genes.
22- line 216 : say explicitly that the reason for not detecting a significant difference in spite of a relatively large effect size is probably related to the low number of genes, conferring low statistical power to detect a difference. An important feature also not highlighted here is that the trend (though not significant) is in the opposite direction than in the buds, and that both the 2-ratio and the free-ratio models agree on these inverted trends. This could be the basis for an interesting comparison.
Thank you for your suggestions.
23- line 220 : needs to explain more clearly how this « free-ratio » differs from the « two-ratio » model.
24- line 232-234 : I don't see why this is necessary. Why not combine both? See also my comment 21 above.
25- line 237 : the «A-model » was not defined before.
26- line 237 : « male-biased » is missing after 343.
27- line 253-258 : briefly explain what these other models are based on and how they are not redundant and instead complement the previous analyses and each other. 28- line 266-268 : the use of a more precise set of codons for male-biased genes than the others (if I understood correctly) could also be interpreted as another sign of stronger selective constraint, no?
Codon usage bias is influenced by many factors, such as levels of gene expression. Highly expressed genes have a stronger codon usage bias and could be encoded by optimal codons for more efficient translation (Frumkin et al., 2018; Parvathy et al., 2022).
29- line 269-291 : removing genes on a post-hoc basis seems statistically suspicious to me. I don't think your analysis has enough power to hand-pick specific categories of genes, and it is not clear what this brings here. I suggest simply removing these analyses and paragraphs.
30- line 325 : say whether this patterns parallels / or not those in animals.
31- line 335 : yes, these biological pieces of information are important and should be given in the introduction already.
32- the discussion should focus at some point on the observation that more femalebiased genes are found in general, but that male-biased genes seem to be under greater selection. How do you reconcile these two apparently contradictory observations?
We found that male-biased genes with high evolutionary rates in male floral buds were associated with functions to abiotic stresses and immune responses (Tables S12 and S13), which suggests that male floral buds through rapidly evolving genes are adapted to mountain climate and the environment in Southwest China compared to female floral buds through high gene expression (lines 387-390).
33- line 355 : not clear how this follows from the previous sentences.
34- line 356-358 : vagiue. not clear what the message of this sentence is.
35- line 378-383 : say that these conclusions rely on the quality of gene annotation in this non-model species, which is probably pretty low (just like any other non-model species).
36- line 403 : this conclusion seems far-fetched. Explain how exactly you reached this conclusion.
37- line 406-416: these speculations on the role of paralogs seem unnecessary, in particular since the de novo transcriptome onto which all analyses are based cannot distinguish orthologs from paralogs.
38- line 417-424. The discussion should not contain new results.
39- line 510 : why were genes with dN/dS >2 discarded here? This might strongly bias the comparison, no? This needs to be properly justified.
40- lines 516-523 : references to the models are missing.
41- line 527: « omega = 1.5 » : why/how was this arbitrary threshold chosen?
42- Fig 2 : write out « buds » and « mature flowers » on top of the graphs
43- Fig 4 : add a phylogeny of the species showing the branch being compared. Also, add the number of genes in each category on each plot.
Thanks, we revised/fixed these issues accordingly.
-

eLife assessment
This valuable paper examines gene expression differences between male and female individuals over the course of flower development in the dioecious angiosperm Trichosantes pilosa. The authors show that male-biased genes evolve faster than female-biased and unbiased genes. This is frequently observed in animals, but this is the first report of such a pattern in plants. In spite of the limited sample size, the evidence is mostly solid and the methods appropriate for a non-model organism. The resources produced will be used by researchers working in the Cucurbitaceae, and the results obtained advance our understanding of the mechanisms of plant sexual reproduction and its evolutionary implications: as such they will broadly appeal to evolutionary biologists and plant biologists.
-

Reviewer #1 (Public Review):
The evolution of dioecy in angiosperms has significant implications for plant reproductive efficiency, adaptation, evolutionary potential, and resilience to environmental changes. Dioecy allows for the specialization and division of labor between male and female plants, where each sex can focus on specific aspects of reproduction and allocate resources accordingly. This division of labor creates an opportunity for sexual selection to act and can drive the evolution of sexual dimorphism.
In the present study, the authors investigate sex-biased gene expression patterns in juvenile and mature dioecious flowers to gain insights into the molecular basis of sexual dimorphism. They find that a large proportion of the plant transcriptome is differentially regulated between males and females with the number of …
Reviewer #1 (Public Review):
The evolution of dioecy in angiosperms has significant implications for plant reproductive efficiency, adaptation, evolutionary potential, and resilience to environmental changes. Dioecy allows for the specialization and division of labor between male and female plants, where each sex can focus on specific aspects of reproduction and allocate resources accordingly. This division of labor creates an opportunity for sexual selection to act and can drive the evolution of sexual dimorphism.
In the present study, the authors investigate sex-biased gene expression patterns in juvenile and mature dioecious flowers to gain insights into the molecular basis of sexual dimorphism. They find that a large proportion of the plant transcriptome is differentially regulated between males and females with the number of sex-biased genes in floral buds being approximately 15 times higher than in mature flowers. The functional analysis of sex-biased genes reveals that chemical defense pathways against herbivores are up-regulated in the female buds along with genes involved in the acquisition of resources such as carbon for fruit and seed production, whereas male buds are enriched in genes related to signaling, inflorescence development and senescence of male flowers. Furthermore, the authors implement sophisticated maximum likelihood methods to understand the forces driving the evolution of sex-biased genes. They highlight the influence of positive and relaxed purifying selection on the evolution of male-biased genes, which show significantly higher rates of non-synonymous to synonymous substitutions than female or unbiased genes. This is the first report (to my knowledge) highlighting the occurrence of this pattern in plants. Overall, this study provides important insights into the genetic basis of sexual dimorphism and the evolution of reproductive genes in Cucurbitaceae.
-

Reviewer #2 (Public Review):
Summary:
This study uses transcriptome sequence from a dioecious plant to compare evolutionary rates between genes with male- and female-biased expression and distinguish between relaxed selection and positive selection as causes for more rapid evolution. These questions have been explored in animals and algae, but few studies have investigated this in dioecious angiosperms, and none have so far identified faster rates of evolution in male-biased genes (though see Hough et al. 2014 https://doi.org/10.1073/pnas.1319227111).
Strengths:
The methods are appropriate to the questions asked. Both the sample size and the depth of sequencing are sufficient, and the methods used to estimate evolutionary rates and the strength of selection are appropriate. The data presented are consistent with faster evolution of …
Reviewer #2 (Public Review):
Summary:
This study uses transcriptome sequence from a dioecious plant to compare evolutionary rates between genes with male- and female-biased expression and distinguish between relaxed selection and positive selection as causes for more rapid evolution. These questions have been explored in animals and algae, but few studies have investigated this in dioecious angiosperms, and none have so far identified faster rates of evolution in male-biased genes (though see Hough et al. 2014 https://doi.org/10.1073/pnas.1319227111).
Strengths:
The methods are appropriate to the questions asked. Both the sample size and the depth of sequencing are sufficient, and the methods used to estimate evolutionary rates and the strength of selection are appropriate. The data presented are consistent with faster evolution of genes with male-biased expression, due to both positive and relaxed selection.
This is a useful contribution to understanding the effect of sex-biased expression in genetic evolution in plants. It demonstrates the range of variation in evolutionary rates and selective mechanisms, and provides further context to connect these patterns to potential explanatory factors in plant diversity such as the age of sex chromosomes and the developmental trajectories of male and female flowers.
Weaknesses:
The presence of sex chromosomes is a potential confounding factor, since there are different evolutionary expectations for X-linked, Y-linked, and autosomal genes. Attempting to distinguish transcripts on the sex chromosomes from autosomal transcripts could provide additional insight into the relative contributions of positive and relaxed selection.
-

Reviewer #3 (Public Review):
The potential for sexual selection and the extent of sexual dimorphism in gene expression have been studied in great detail in animals, but hardly examined in plants so far. In this context, the study by Zhao, Zhou et al. al represents a welcome addition to the literature.
Relative to the previous studies in Angiosperms, the dataset is interesting in that it focuses on reproductive rather than somatic tissues (which makes sense to investigate sexual selection), and includes more than a single developmental stage (buds + mature flowers).
Some aspects of the presentation have been improved in this new version of the manuscript. Specifically:
- the link between sex-biased and tissue-biased genes is now slightly clearer,
- the limitation related to the de novo assembled transcriptome is now formally …Reviewer #3 (Public Review):
The potential for sexual selection and the extent of sexual dimorphism in gene expression have been studied in great detail in animals, but hardly examined in plants so far. In this context, the study by Zhao, Zhou et al. al represents a welcome addition to the literature.
Relative to the previous studies in Angiosperms, the dataset is interesting in that it focuses on reproductive rather than somatic tissues (which makes sense to investigate sexual selection), and includes more than a single developmental stage (buds + mature flowers).
Some aspects of the presentation have been improved in this new version of the manuscript. Specifically:
- the link between sex-biased and tissue-biased genes is now slightly clearer,
- the limitation related to the de novo assembled transcriptome is now formally acknowledged,
- the interpretation of functional categories of the genes identified is more precise,
- the legends of supplementary figures have been improved
- a large number of typos have been fixed.However, overall the analyses are largely unchanged and the manuscript did not mature much in response to this first round of reviews. As I detail below, many of the relevant and constructive suggestions by the previous reviewers were not taken into account in this revision. For instance:
- Reviewer 2 made precise suggestions for trying to take into account the potential confonding factor of sex-chromosomes. This suggestion was not followed.
- Reviewer 1 & 3 indicated that results were mentioned in the discussion section without having been described before. This was not fixed in this new version.
- Reviewer 1 asked for a comparison between the number of de novo assembled unigenes in this transcriptome and the number of genes in other Cucurbitaceae species. I could not see this comparison reported.
- Reviewer 1 pointed out that permutation tests were more appropriate, but no change was made to the manuscript.
- Reviewer 3 pointed out the small sample size (both for the RNA-seq and the phylogenetic analysis), but again this limitation is not acknowledged very clearly.
- Reviewer 1 & 3 pointed out that Fig 3 was hard to understand and asked for clarifications that I did not see in the text and the figure in unchanged.
- Reviewer 3 suggested to combine all genes with sex-bias expression when evaluating the evolutionary rate, in addition to the analyses already done. This suggestion was not followed.
- Reviewer 3 pointed out that hand-picking specific categories of genes was not statistically valid, and in fact not necessary in the present context. This was not changed.
- Reviewer 1 asked for all data to be public, but I could not find in the manuscript where the link to the data on ResearchGate was provided.
- Reviewers 1 & 3 pointed out that since only two tissues were compared, the claims on pleiotropy should have been toned down, but no change was made to the text.
- Reviewer 1 asked for a clarification on which genes are plotted on the heatmap of Fig3C and an explanation of the color scale. No change was made.
- Reviewer 1 asked for panel B in Fig 5 and 6 to be removed. They are still there. They asked for abbreviations to be explained in the legend of Fig S8. This was not done. They asked for details about coluln headers. Such detailed were not added. They asked for more recent references on line 53-56 : this was not done. -

-

eLife assessment
This valuable paper examines gene expression differences between male and female individuals over the course of flower development in the dioecious angiosperm Trichosantes pilosa. The authors show that male-biased genes evolve faster than female-biased and unbiased genes, which is frequently observed in animals but this is the first report of such a pattern in plants. In spite of the limited sample size, the reviewers found the evidence to be mostly solid and the methods appropriate for a non-model organism. The resources produced will be used by researchers working in the Cucurbitaceae, and the results obtained advance our understanding of the mechanisms of plant sexual reproduction and its evolutionary implications: as such they will broadly appeal to evolutionary biologists and plant biologists.
-

Reviewer #1 (Public Review):
The evolution of dioecy in angiosperms has significant implications for plant reproductive efficiency, adaptation, evolutionary potential, and resilience to environmental changes. Dioecy allows for the specialization and division of labor between male and female plants, where each sex can focus on specific aspects of reproduction and allocate resources accordingly. This division of labor creates an opportunity for sexual selection to act and can drive the evolution of sexual dimorphism.
In the present study, the authors investigate sex-biased gene expression patterns in juvenile and mature dioecious flowers to gain insights into the molecular basis of sexual dimorphism. They find that a large proportion of the plant transcriptome is differentially regulated between males and females with the number of …
Reviewer #1 (Public Review):
The evolution of dioecy in angiosperms has significant implications for plant reproductive efficiency, adaptation, evolutionary potential, and resilience to environmental changes. Dioecy allows for the specialization and division of labor between male and female plants, where each sex can focus on specific aspects of reproduction and allocate resources accordingly. This division of labor creates an opportunity for sexual selection to act and can drive the evolution of sexual dimorphism.
In the present study, the authors investigate sex-biased gene expression patterns in juvenile and mature dioecious flowers to gain insights into the molecular basis of sexual dimorphism. They find that a large proportion of the plant transcriptome is differentially regulated between males and females with the number of sex-biased genes in floral buds being approximately 15 times higher than in mature flowers. The functional analysis of sex-biased genes reveals that chemical defense pathways against herbivores are up-regulated in the female buds along with genes involved in the acquisition of resources such as carbon for fruit and seed production, whereas male buds are enriched in genes related to signaling, inflorescence development and senescence of male flowers. Furthermore, the authors implement sophisticated maximum likelihood methods to understand the forces driving the evolution of sex-biased genes. They highlight the influence of positive and relaxed purifying selection on the evolution of male-biased genes, which show significantly higher rates of non-synonymous to synonymous substitutions than female or unbiased genes. This is the first report (to my knowledge) highlighting the occurrence of this pattern in plants. Overall, this study provides important insights into the genetic basis of sexual dimorphism and the evolution of reproductive genes in Cucurbitaceae.
There are, however, parts of the manuscript that are not clearly described or could be otherwise improved.
- The number of denovo-assembled unigenes seems large and I would like to know how it compares to the number of genes in other Cucurbitaceae species. The presence of alternatively assembled isoforms or assembly artifacts may be still high in the final assembly and inflate the numbers of identified sex-biased genes.
- It is interesting that the majority of sex-biased genes are present in the floral buds but not in the mature flowers. I think this pattern could be explored in more detail, by investigating the expression of male and female sex-biased genes throughout the flower development in the opposite sex. It is also not clear how the expression of the sex-biased genes found in the buds changes when buds and mature flowers are compared within each sex.
- The statistical analysis of evolutionary rates between male-biased, female-biased, and unbiased genes is performed on samples with very different numbers of observations, therefore, a permutation test seems more appropriate here.
- The impact of pleiotropy on the evolutionary rates of male-biased genes is speculative since only two tissue samples (buds and mature flowers) are used. More tissue types need to be included to draw any meaningful conclusions here.
-

Reviewer #2 (Public Review):
Summary:
This study uses transcriptome sequence from a dioecious plant to compare evolutionary rates between genes with male- and female-biased expression and distinguish between relaxed selection and positive selection as causes for more rapid evolution. These questions have been explored in animals and algae, but few studies have investigated this in dioecious angiosperms, and none have so far identified faster rates of evolution in male-biased genes (though see Hough et al. 2014 https://doi.org/10.1073/pnas.1319227111).Strengths:
The methods are appropriate to the questions asked. Both the sample size and the depth of sequencing are sufficient, and the methods used to estimate evolutionary rates and the strength of selection are appropriate. The data presented are consistent with faster evolution of …Reviewer #2 (Public Review):
Summary:
This study uses transcriptome sequence from a dioecious plant to compare evolutionary rates between genes with male- and female-biased expression and distinguish between relaxed selection and positive selection as causes for more rapid evolution. These questions have been explored in animals and algae, but few studies have investigated this in dioecious angiosperms, and none have so far identified faster rates of evolution in male-biased genes (though see Hough et al. 2014 https://doi.org/10.1073/pnas.1319227111).Strengths:
The methods are appropriate to the questions asked. Both the sample size and the depth of sequencing are sufficient, and the methods used to estimate evolutionary rates and the strength of selection are appropriate. The data presented are consistent with faster evolution of genes with male-biased expression, due to both positive and relaxed selection.This is a useful contribution to understanding the effect of sex-biased expression in genetic evolution in plants. It demonstrates the range of variation in evolutionary rates and selective mechanisms, and provides further context to connect these patterns to potential explanatory factors in plant diversity such as the age of sex chromosomes and the developmental trajectories of male and female flowers.
Weaknesses:
The presence of sex chromosomes is a potential confounding factor, since there are different evolutionary expectations for X-linked, Y-linked, and autosomal genes. Attempting to distinguish transcripts on the sex chromosomes from autosomal transcripts could provide additional insight into the relative contributions of positive and relaxed selection. -

Reviewer #3 (Public Review):
The potential for sexual selection and the extent of sexual dimorphism in gene expression have been studied in great detail in animals, but hardly examined in plants so far. In this context, the study by Zhao, Zhou et al. al represents a welcome addition to the literature.
Relative to the previous studies in Angiosperms, the dataset is interesting in that it focuses on reproductive rather than somatic tissues (which makes sense to investigate sexual selection), and includes more than a single developmental stage (buds + mature flowers).
The main limitation of the study is the very low number of samples analyzed, with only three replicate individuals per sex (i.e. the whole study is built on six individuals only). This provides low power to detect differential expression. Along the same line, only three …
Reviewer #3 (Public Review):
The potential for sexual selection and the extent of sexual dimorphism in gene expression have been studied in great detail in animals, but hardly examined in plants so far. In this context, the study by Zhao, Zhou et al. al represents a welcome addition to the literature.
Relative to the previous studies in Angiosperms, the dataset is interesting in that it focuses on reproductive rather than somatic tissues (which makes sense to investigate sexual selection), and includes more than a single developmental stage (buds + mature flowers).
The main limitation of the study is the very low number of samples analyzed, with only three replicate individuals per sex (i.e. the whole study is built on six individuals only). This provides low power to detect differential expression. Along the same line, only three species were used to evaluate the rates of non-synonymous to synonymous substitutions, which also represents a very limited dataset, in particular when trying to fit parameter-rich models such as those implemented here.
A third limitation relates to the absence of a reference genome for the species, making the use of a de novo transcriptome assembly necessary, which is likely to lead to a large number of incorrectly assembled transcripts. Of course, the production of a reference transcriptome in this non-model species is already a useful resource, but this point should at least be acknowledged somewhere in the manuscript.
Each of these shortcomings is relatively important, and together they strongly limit the scope of the conclusions that can be made, and they should at least be acknowledged more prominently. The study is valuable in spite of these limitations and the topic remains grossly understudied, so I think the study will be of interest to researchers in the field, and hopefully inspire further, more comprehensive analyses.
-