The target of rapamycin signaling pathway regulates vegetative development, aflatoxin biosynthesis, and pathogenicity in Aspergillus flavus
Curation statements for this article:-
Curated by eLife
eLife assessment
This manuscript provides important information about the influence of TOR signaling pathway on development and aflatoxin production in the plant and human fungal pathogen Aspergillus flavus. Compared to an earlier version, the authors have addressed most of the concerns of the reviewers, including the convincing demonstration of the essential TOR pathway in this fungus by constructing a xylose promoter mutant strain.
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
The target of rapamycin (TOR) signaling pathway is highly conserved and plays a crucial role in diverse biological processes in eukaryotes. Despite its significance, the underlying mechanism of the TOR pathway in Aspergillus flavus remains elusive. In this study, we comprehensively analyzed the TOR signaling pathway in A. flavus by identifying and characterizing nine genes that encode distinct components of this pathway. The FK506-binding protein Fkbp3 and its lysine succinylation are important for aflatoxin production and rapamycin resistance. The TorA kinase plays a pivotal role in the regulation of growth, spore production, aflatoxin biosynthesis, and responses to rapamycin and cell membrane stress. As a significant downstream effector molecule of the TorA kinase, the Sch9 kinase regulates aflatoxin B 1 (AFB 1 ) synthesis, osmotic and calcium stress response in A. flavus, and this regulation is mediated through its S_TKc, S_TK_X domains, and the ATP-binding site at K340. We also showed that the Sch9 kinase may have a regulatory impact on the high osmolarity glycerol (HOG) signaling pathway. TapA and TipA, the other downstream components of the TorA kinase, play a significant role in regulating cell wall stress response in A. flavus . Moreover, the members of the TapA-phosphatase complexes, SitA and Ppg1, are important for various biological processes in A. flavus , including vegetative growth, sclerotia formation, AFB 1 biosynthesis, and pathogenicity. We also demonstrated that SitA and Ppg1 are involved in regulating lipid droplets (LDs) biogenesis and cell wall integrity (CWI) signaling pathways. In addition, another phosphatase complex, Nem1/Spo7, plays critical roles in hyphal development, conidiation, aflatoxin production, and LDs biogenesis. Collectively, our study has provided important insight into the regulatory network of the TOR signaling pathway and has elucidated the underlying molecular mechanisms of aflatoxin biosynthesis in A. flavus .
Article activity feed
-

-

-

Author response:
The following is the authors’ response to the previous reviews.
Reviewer #1 (Public Review):
While I acknowledge the authors' effort in conducting Southern blot analysis to address my prior concern regarding the presence of dual copies of torA and tapA, I find their current resolution inadequate. Specifically, the simple deletion of the respective result sections for torA and tapA significantly impacts the overall significance of this study. The repeated unsuccessful attempts to generate correct mutants only offer circumstantial evidence, as technical issues may have been a contributing factor. Therefore, instead of merely removing these sections, it is essential for the authors to present more compelling experimental data demonstrating that torA and tapA are indeed vital for the viability of A. flavus. Such data would …
Author response:
The following is the authors’ response to the previous reviews.
Reviewer #1 (Public Review):
While I acknowledge the authors' effort in conducting Southern blot analysis to address my prior concern regarding the presence of dual copies of torA and tapA, I find their current resolution inadequate. Specifically, the simple deletion of the respective result sections for torA and tapA significantly impacts the overall significance of this study. The repeated unsuccessful attempts to generate correct mutants only offer circumstantial evidence, as technical issues may have been a contributing factor. Therefore, instead of merely removing these sections, it is essential for the authors to present more compelling experimental data demonstrating that torA and tapA are indeed vital for the viability of A. flavus. Such data would enhance the overall significance of this study.
We agree and appreciate reviewer's important comments on our manuscript. In this version, we address this issue by providing additional experimental data to further support the importance of torA and tapA in the viability of A. flavus. We conducted additional experiments to generate more compelling evidence regarding the essential role of torA and tapA in the growth and development of A. flavus. We constructed a mutant strain (xylPtorA) using an xylose-inducible promoter, which allows for conditional induction with the addition of xylose (Lines 204-238, page 10).
Due to the unsuccessful construction of TapA knockout strains and xylose promoter replacement strains, we used homologous recombination to replace the original promoter with the gpdA strong promoter for overexpression of tapA (OE::tapA). We thank reviewer for highlighting this important aspect, and we revise our manuscript accordingly to enhance its overall significance (Lines 277-297, page 13). We are grateful for the opportunity to enhance our manuscript and believe these revisions provide a more comprehensive understanding of the roles of torA and tapA in A. flavus.
Reviewer #1 (Recommendations For The Authors):
Minor comments
Lines 421-423 and 465-466: these sentences are grammatically awkward. Please rephrase them.
Thank you for your feedback on our manuscript. We conducted additional experiments, so we have removed the sentence from the manuscript to maintain coherence and avoid redundancy.
Reviewer #2 (Public Review):
In this study, authors identified TOR, HOG and CWI signaling network genes as modulators of the development, aflatoxin biosynthesis and pathogenicity of A. flavus by gene deletions combined with phenotypic observation. They also analyzed the specific regulatory process and proposed that the TOR signaling pathway interacts with other signaling pathways (MAPK, CWI, calcineurin-CrzA pathway) to regulate the responses to various environmental stresses. Notably, they found that FKBP3 is involved in sclerotia and aflatoxin biosynthesis and rapamycin resistance in A. flavus, especially that the conserved site K19 of FKBP3 plays a key role in regulating aflatoxin biosynthesis. In general, the study involved a heavy workload and the findings are potentially interesting and important for understanding or controlling the aflatoxin biosynthesis. However, the findings have not been deeply explored and the conclusions mostly are based on parallel phenotypic observations.
Thank you for your constructive comments on our manuscript. In response to your comments, we have conducted additional experiments, including the construction of a xylose promoter mutant strain and an overexpression strain. We have also expanded the discussion section to provide a more comprehensive analysis of our findings in the context of existing literature. Thank you again for your insightful feedback, which has been instrumental in improving the quality of our work. (Lines 464-469, page 22).
Reviewer #2 (Recommendations For The Authors):
Point 1: Our findings revealed that both the tor and tapA genes are present in double copies in our strains, which guided our decision to construct single-copy deletion strains using homologous recombination However, the tor gene in A. flavus exhibited varying copy numbers, as was confirmed by absolute quantification PCR at the genome level (Table S1). However, it is hard to understand for Table S1: Estimation of copy number of tor gene in A. flavus toro and sumoo stand for the initial copy number, and the data are graphed as the mean {plus minus} 95%confidence limit. CN is copy number. As indicated in the Methods, Using sumo gene as reference, the tor and tapA gene copy number was calculated by standard curve. In Table S1 of WT, for tor gene, CN value is1412537 compared to 1698243 in tor+/-, for the reference gene sumo,794328 compared to1584893, how these data could support copy gene numbers of tor?
Thank you for your insightful comments. We understand the confusion with the data presented in Table S1 regarding the copy number estimation of the torA gene in A. flavus. We apologize for not providing a clear explanation for the data in the table. Quantitative real-time PCR (qPCR) is widely used to determine the copy number of a specific gene. It involves amplifying the gene of interest and a reference gene simultaneously using specific primers and probes. By comparing the amplification curves of the gene of interest and the reference gene, we can estimate the relative copy number of the gene.
To address your concern and provide more accurate information, we have re-performed the copy number analysis using southern blot. Southern blot analysis allows for the direct estimation of gene copy number by hybridizing genomic DNA with a specific probe for the gene. This method provides more reliable and accurate results in determining gene copy numbers. We discovered that the A. flavus genome contains a single copy of the torA gene. Consequently, we conducted additional experiments to elucidate its function. Specifically, we generated strains with a xylose-inducible promoter system to modulate the expression of torA (Lines 204-238, page 10).
Point 2: In response: For the knockout of the FRB domain, we used the homologous recombination method, but because tor genes are double-copy genes, there are also double copies in the FRB domain. Despite our efforts, we encountered challenges in precisely determining the location of the other copy of the tor gene. I could not understand these consistent data, why not for using sequencing?
Thank you for your valuable feedback. We determined again and confirmed that the torA gene is a single copy. So we removed this part of the results to avoid any ambiguity or potential misinterpretation.
Point 3: Response in Due to the large number of genes involved, we did not perform a complementation experiment. If there were no complementation data, how to demonstrate data are solid?
Thank you for your important suggestion. We understand that complementation experiments are commonly used to validate gene deletions. Therefore, to ensure the reliability of our data, we have conducted supplementary experiments on specific gene deletions, such as ΔsitA-C and Δppg1-C. Thank you again for your positive comments and valuable suggestions, which have significantly contributed to enhancing the quality of our manuscript (Lines 320-322, page 15).
Point 4: Acknowledge the confusion? We acknowledge the confusion in our presentation and will ensure that accurate genetic nomenclature is used consistently
Thank you for your comments on our manuscript. We recognize the importance of precise and consistent use of genetic nomenclature, as it is critical for the clarity and integrity of our research findings. We have carefully reviewed the sections of our manuscript where genetic terms were used and have made the necessary corrections to ensure that all nomenclature is accurate and used consistently throughout the text.
Point 5: In the revised version of new manuscript, southern blotting was carried out and found only one copy was existed for tested genes at last. Thus, whole manuscript conclusions should be changed. In addition, Reviewer 1 suggestion for using Illumina-sequence strategy, their tor and tapA mutants could be verified whether they are aneuploid?
We would like to express our gratitude for your insightful comments and suggestions. Following the new experimental data obtained from Southern blotting, we have identified that only one copy of the tested genes exists, and we have revised our conclusions throughout the manuscript. This has led to a significant reinterpretation of our results and a reassessment of the implications for our study. Based on this result, we designed and constructed strains with the tor gene under the control of a xylose-inducible promoter. This approach allows for the conditional expression of the tor gene. Thank you once again for your meticulous review (Lines 204-238, page 10).
-

eLife assessment
This manuscript provides important information about the influence of TOR signaling pathway on development and aflatoxin production in the plant and human fungal pathogen Aspergillus flavus. Compared to an earlier version, the authors have addressed most of the concerns of the reviewers, including the convincing demonstration of the essential TOR pathway in this fungus by constructing a xylose promoter mutant strain.
-

Reviewer #1 (Public Review):
This paper reports the useful discovery of the roles and signaling components of the TOR pathway in vegetative growth, sexual development, stress response, and aflatoxin production in Aspergillus flavus.
While I acknowledge the authors' effort in conducting Southern blot analysis to address my prior concern regarding the presence of dual copies of torA and tapA, I find their current resolution inadequate. Specifically, the simple deletion of the respective result sections for torA and tapA significantly impacts the overall significance of this study. The repeated unsuccessful attempts to generate correct mutants only offer circumstantial evidence, as technical issues may have been a contributing factor. Therefore, instead of merely removing these sections, it is essential for the authors to present more …
Reviewer #1 (Public Review):
This paper reports the useful discovery of the roles and signaling components of the TOR pathway in vegetative growth, sexual development, stress response, and aflatoxin production in Aspergillus flavus.
While I acknowledge the authors' effort in conducting Southern blot analysis to address my prior concern regarding the presence of dual copies of torA and tapA, I find their current resolution inadequate. Specifically, the simple deletion of the respective result sections for torA and tapA significantly impacts the overall significance of this study. The repeated unsuccessful attempts to generate correct mutants only offer circumstantial evidence, as technical issues may have been a contributing factor. Therefore, instead of merely removing these sections, it is essential for the authors to present more compelling experimental data demonstrating that torA and tapA are indeed vital for the viability of A. flavus. Such data would enhance the overall significance of this study.
-

Reviewer #2 (Public Review):
In this study, authors identified TOR, HOG and CWI signaling network genes as modulators of the development, aflatoxin biosynthesis and pathogenicity of A. flavus by gene deletions combined with phenotypic observation. They also analyzed the specific regulatory process and proposed that the TOR signaling pathway interacts with other signaling pathways (MAPK, CWI, calcineurin-CrzA pathway) to regulate the responses to various environmental stresses. Notably, they found that FKBP3 is involved in sclerotia and aflatoxin biosynthesis and rapamycin resistance in A. flavus, especially that the conserved site K19 of FKBP3 plays a key role in regulating aflatoxin biosynthesis. In general, the study involved a heavy workload and the findings are potentially interesting and important for understanding or controlling …
Reviewer #2 (Public Review):
In this study, authors identified TOR, HOG and CWI signaling network genes as modulators of the development, aflatoxin biosynthesis and pathogenicity of A. flavus by gene deletions combined with phenotypic observation. They also analyzed the specific regulatory process and proposed that the TOR signaling pathway interacts with other signaling pathways (MAPK, CWI, calcineurin-CrzA pathway) to regulate the responses to various environmental stresses. Notably, they found that FKBP3 is involved in sclerotia and aflatoxin biosynthesis and rapamycin resistance in A. flavus, especially that the conserved site K19 of FKBP3 plays a key role in regulating aflatoxin biosynthesis. In general, the study involved a heavy workload and the findings are potentially interesting and important for understanding or controlling the aflatoxin biosynthesis. However, the findings have not been deeply explored and the conclusions mostly are based on parallel phenotypic observations.
-

-

Author Response
The following is the authors’ response to the original reviews.
Response to Reviewer 1 Comments (PublicReview)
Point 1: First, the authors should provide more convincing data showing that tor and tapA genes are indeed duplicated genes in A. flavus. The authors appeared to use the A. flavus PTS strain as a parental strain for constructing the tor and tapA mutants. If so, the A. flavus CA14 strain (Hua et al., 2007) should be the parental wild-type strain for the A. flavus PTS strain. I did a BLAST search in NCBI for the torA (AFLA_044350) and tapA (AFLA_092770) genes using the most recent CA14 genome assembly sequence (GCA_014784225.2) and only found one allele for each gene: torA on chromosome 7 and tapA on chromosome 3. I could not find any other parts with similar sequences. Even in another popular A. flavus …
Author Response
The following is the authors’ response to the original reviews.
Response to Reviewer 1 Comments (PublicReview)
Point 1: First, the authors should provide more convincing data showing that tor and tapA genes are indeed duplicated genes in A. flavus. The authors appeared to use the A. flavus PTS strain as a parental strain for constructing the tor and tapA mutants. If so, the A. flavus CA14 strain (Hua et al., 2007) should be the parental wild-type strain for the A. flavus PTS strain. I did a BLAST search in NCBI for the torA (AFLA_044350) and tapA (AFLA_092770) genes using the most recent CA14 genome assembly sequence (GCA_014784225.2) and only found one allele for each gene: torA on chromosome 7 and tapA on chromosome 3. I could not find any other parts with similar sequences. Even in another popular A. flavus wild-type strain, NRRL3357, both torA and tapA exist as a single allele. Based on the published genome assembly data for A. flavus, there is no evidence to support the idea that tor and tapA exist as copies of each other. Therefore, the authors could perform a Southern blot analysis to further verify their claim. If torA and tapA indeed exist as duplicate copies in different chromosomal locations, Southern blot data could provide supporting results.
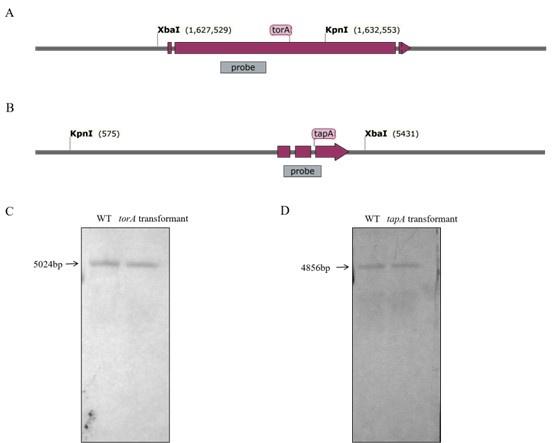
Response 1: We thank the reviewer for their insightful observation. Based on the southern blot analysis results presented in Figure 1, we have determined that torA and tapA are single-copy genes. Additionally, we conducted protoplast transformation experiments repeated several times. which revealed that both torA and tapA transformants exhibited ectopic mutations. It is plausible that the deletion of torA and tapA genes may lead to the demise of A. flavus, this phenomenon is consistent with previous studies conducted on the fungus Fusarium graminearum[1].To ensure the rigor of the study, we have retracted the previously incorrect conclusion. We once again express our heartfelt appreciation to the experts for their valuable suggestions.
Author response image 1.
Fig.1 Southern blot hybridization analyses of WT, torA, and tapA transformants. (A) The structure diagram of the torA gene. (B) The structure diagram of the tapA gene. (C) Southern blot hybridization analyses of torA gene. (D) Southern blot hybridization analyses of tapA gene.
Point 2: Second, the authors should consider the possibility of aneuploidy for their constructed mutants. When an essential gene is targeted for deletion, aneuploidy often occurs even in a fungal strain without the "ku" mutation, which results in seemingly dual copies of the gene. As the authors appear to use the A. flavus PTS strain having the "ku" mutation, the parental strain has increased genome instability, which may result in enhanced chromosomal rearrangements. So, it will be necessary to Illumina-sequence their tor and tapA mutants to make sure that they are not aneuploidy.
Response 2: Thank you for your comment. Based on the sequencing results of the torA and tapA mutants, it was determined that the torA and tapA genes were still present in both mutants. In this case, it suggests that the torA and tapA genes may have undergone a genetic rearrangement or insertion at a different site in the mutant strains.
Point 3: Furthermore, the genetic nomenclature +/- and -/- should be reserved for heterozygous and homozygous mutants in a diploid strain. As A. flavus is not a diploid strain, this type of description could cause confusion for the readers.
Response 3: Thank you for your suggestion. We acknowledge your concerns about potential confusion caused by using this type of description, and we agree that it is best to avoid any misunderstandings for readers. Therefore, we have decided to remove this part of the content from the manuscript.
Response to Reviewer 2 Comments (PublicReview)
Point 1: However, findings have not been deeply explored and conclusions are mostly are based on parallel phenotypic observations. In addition, there are some concerns for the conclusions.
Response 1: We are grateful for the suggestion. We conduct additional experiments and analyses to provide a more comprehensive understanding and address concerns raised.
Response to Reviewer 3 Comments (PublicReview)
Point 1: The paper by Li et al. describes the role of the TOR pathway in Aspergillus flavus. The authors tested the effect of rapamycin in WT and different deletion strains. This paper is based on a lot of experiments and work but remains rather descriptive and confirms the results obtained in other fungi. It shows that the TOR pathway is involved in conidiation, aflatoxin production, pathogenicity, and hyphal growth. This is inferred from rapamycin treatment and TOR1/2 deletions. Rapamycin treatment also causes lipid accumulation in hyphae. The phenotypes are not surprising as they have been shown already for several fungi. In addition, one caveat is in my opinion that the strains grow very slowly and this could cause many downstream effects. Several kinases and phosphatases are involved in the TOR pathway. They were known from S. cerevisiae or filamentous fungi. The authors characterized them as well with knock-out approaches.
Response 1: Thank you for your comment. The role of the target of rapamycin (TOR) signaling pathway is of fundamental importance in the physiological processes of diverse eukaryotic organisms. Nevertheless, its precise involvement in regulating the developmental and virulent characteristics of opportunistic pathogenic fungi, such as A. flavus, has yet to be fully elucidated. Furthermore, the mechanistic underpinnings of TOR pathway activity specifically in A. flavus remain largely unresolved. Consequently, our study represents a significant contribution as the first comprehensive exploration of the conserved TOR signaling pathway encompassing a majority of its constituent genes in A. flavus.
Response to Reviewer 1 Comments (Recommendations For The Authors)
Point 1: In Table S3, the authors indicated that the Δku70 ΔniaD ΔpyrG::pyrG strain is A. flavus wild-type strain. However, this strain is not a wild-type strain because it seems like a control strain after introducing the pyrG gene into the A. flavus PTS strain (Δku70 ΔniaD ΔpyrG). So please indicate the real wild-type A. flavus strain name to help readers find out its original genome sequence data. Also, the reference for this Δku70 ΔniaD ΔpyrG::pyrG strain is "saved in our lab". This is not an eligible reference. If you use this control strain for the first time in this study, it should be described as "In this study". Otherwise, please indicate the proper reference for which the strain was first used.
Response 1: Thank you for your valuable feedback on our manuscript. We appreciate your attention to detail and the opportunity to clarify the information regarding the strain in Table S3. The A. flavus CA14 strain which produces aflatoxins and large sclerotia was isolated from a pistachio bud in the Wolfskill Grant Experimental Farm (University of Davis, Winters, California, USA)[2]. The A. flavus CA14 strain is the parental wild-type strain for the A. flavus CA14 PTs (Δku70, ΔniaD, ΔpyrG) strain. The recipient strain CA14 PTs has been used satisfactorily in gene knockout and subsequent genetic complementation experiments[3]. In this study, the A. flavus CA14 PTs strain was used as the transformation recipient strain, and the control strain (Δku70, ΔniaD, ΔpyrG::pyrG) created by introducing the pyrG gene into the A. flavus CA14 PTs strain. Refer to previously published literature[4],this control strain (Δku70, ΔniaD, ΔpyrG::pyrG) was named wild-type strain. Therefore, this control strain was also named wild-type strain in this study. As this control strain is indeed used in this study, we will revise the reference to "In this study" Once again, we appreciate your keen attention to detail and thank you for bringing these issues to our attention.
Response to Reviewer 2 Comments (Recommendations For The Authors)
Point 1: As in response: However, the tor gene in A. flavus exhibited varying copy numbers, as was confirmed by absolute quantification PCR at the genome level (Table S1). However, it is hard to understand Table S1: Estimation of copy number of tor gene in A. flavus toro and sumoo stand for the initial copy number, and the data are figured as the mean {plus minus} 95% confidence limit. CN is copy number. As indicated in the section of Method, using sumo gene as reference, the tor and tapA gene copy number was calculated by standard curve. In Table S1 of WT, for tor gene, CN value is 1412537 compared to 1698243 in tor+/-, for the reference gene sumo,794328 compared to1584893, how these data could support copy gene numbers of tor?
Response 1: Thank you for your suggestion. We understand the confusion with the data presented in Table S1 regarding the copy number estimation of the tor gene in A. flavus. We apologize for not providing a clear explanation for the data in the table. Quantitative real-time PCR (qPCR) is widely used to determine the copy number of a specific gene. It involves amplifying the gene of interest and a reference gene simultaneously using specific primers and probes. By comparing the amplification curves of the gene of interest and the reference gene, you can estimate the relative copy number of the gene.
To address your concern and provide more accurate information, we have re-performed the copy number analysis using southern blot. Southern blot analysis allows for the direct estimation of gene copy number by hybridizing genomic DNA with a specific probe for the gene. This method provides more reliable and accurate results in determining gene copy numbers. The southern blot analysis results are presented in Figure 1.
We appreciate your input and apologize for any confusion caused by the earlier presentation of the data.
Point 2: In response: For the knockout of the FRB domain, we used the homologous recombination method, but because tor genes are double-copy genes, there are also double copies in the FRB domain. Despite our efforts, we encountered challenges in precisely determining the location of the other copy of the tor gene. I could not understand these consistent data, why not for using sequencing?
Response 2: Thank you for your comment. We observed that the torA gene is a single copy. We removed this part of the results to avoid any ambiguity or potential misinterpretation.
Point 3: Response in Due to the large number of genes involved, we did not perform a complementation experiment. If there were no complementation data, how to demonstrate data are solid?
Response 3: Thank you for your important suggestion. We understand that complementation experiments are commonly used to validate gene deletions. Therefore, to ensure the reliability of our data, we have conducted supplementary experiments on specific gene deletions, such as ΔsitA-C and Δppg1-C. Thank you again for your positive comments and valuable suggestions to improve the quality of our manuscript.
References:
(1) Yu F, Gu Q, Yun Y, et al. The TOR signaling pathway regulates vegetative development and virulence in Fusarium graminearum. New Phytol. 2014; 203(1): 219-32.
(2) Hua SS, Tarun AS, Pandey SN, Chang L, Chang PK. Characterization of AFLAV, a Tf1/Sushi retrotransposon from Aspergillus flavus. Mycopathologia. 2007 Feb;163(2):97-104.
(3) Chang PK, Scharfenstein LL, Mack B, Hua SST. Genome sequence of an Aspergillus flavus CA14 strain that is widely used in gene function studies. Microbiol Resour Announc. 2019 Aug 15;8(33):e00837-19.
(4) Zhu Z, Yang M, Yang G, Zhang B, Cao X, Yuan J, Ge F, Wang S. PP2C phosphatases Ptc1 and Ptc2 dephosphorylate PGK1 to regulate autophagy and aflatoxin synthesis in the pathogenic fungus Aspergillus flavus. mBio. 2023 Oct 31;14(5):e0097723.
-

eLife assessment
This important study presents relevant information about the involvement of TOR pathway in aflatoxin production by Aspergillus flavus. However, some of the presentation is confusing, leaving the study in its current form is incomplete. The strength of the evidence could be augmented with additional experiments and reorganization of the manuscript aiming to fully understand and characterize the involvement of TOR pathway in A. flavus aflatoxin production.
-

Reviewer #1 (Public Review):
While I acknowledge the authors' effort in conducting Southern blot analysis to address my prior concern regarding the presence of dual copies of torA and tapA, I find their current resolution inadequate. Specifically, the simple deletion of the respective result sections for torA and tapA significantly impacts the overall significance of this study. The repeated unsuccessful attempts to generate correct mutants only offer circumstantial evidence, as technical issues may have been a contributing factor. Therefore, instead of merely removing these sections, it is essential for the authors to present more compelling experimental data demonstrating that torA and tapA are indeed vital for the viability of A. flavus. Such data would enhance the overall significance of this study.
-

Reviewer #2 (Public Review):
In this study, authors identified TOR, HOG and CWI signaling network genes as modulators of the development, aflatoxin biosynthesis and pathogenicity of A. flavus by gene deletions combined with phenotypic observation. They also analyzed the specific regulatory process and proposed that the TOR signaling pathway interacts with other signaling pathways (MAPK, CWI, calcineurin-CrzA pathway) to regulate the responses to various environmental stresses. Notably, they found that FKBP3 is involved in sclerotia and aflatoxin biosynthesis and rapamycin resistance in A. flavus, especially that the conserved site K19 of FKBP3 plays a key role in regulating aflatoxin biosynthesis. In general, the study involved a heavy workload and the findings are potentially interesting and important for understanding or controlling …
Reviewer #2 (Public Review):
In this study, authors identified TOR, HOG and CWI signaling network genes as modulators of the development, aflatoxin biosynthesis and pathogenicity of A. flavus by gene deletions combined with phenotypic observation. They also analyzed the specific regulatory process and proposed that the TOR signaling pathway interacts with other signaling pathways (MAPK, CWI, calcineurin-CrzA pathway) to regulate the responses to various environmental stresses. Notably, they found that FKBP3 is involved in sclerotia and aflatoxin biosynthesis and rapamycin resistance in A. flavus, especially that the conserved site K19 of FKBP3 plays a key role in regulating aflatoxin biosynthesis. In general, the study involved a heavy workload and the findings are potentially interesting and important for understanding or controlling the aflatoxin biosynthesis. However, the findings have not been deeply explored and the conclusions mostly are based on parallel phenotypic observations.
-

-

Author Response
The following is the authors’ response to the original reviews.
Response to Reviewer 1 Comments (Public Review):
Point 1: While the authors provided a large amount of data regarding the genes involved in the TOR pathway, it is highly descriptive and mostly confirmative data, as numerous papers have already shown that the TOR pathway plays essential roles in a myriad of biological processes in multiple fungi.
Response 1: Thank you for your comment. The target of rapamycin (TOR) signal pathway plays critical roles in various eukaryotic organisms. However, its specific role in controlling the development and virulence of opportunistic pathogenic fungi like A. flavus has remained unclear. Additionally, the underlying mechanism of the TOR pathway remains elusive in the A. flavus. As such, our study provides a useful …
Author Response
The following is the authors’ response to the original reviews.
Response to Reviewer 1 Comments (Public Review):
Point 1: While the authors provided a large amount of data regarding the genes involved in the TOR pathway, it is highly descriptive and mostly confirmative data, as numerous papers have already shown that the TOR pathway plays essential roles in a myriad of biological processes in multiple fungi.
Response 1: Thank you for your comment. The target of rapamycin (TOR) signal pathway plays critical roles in various eukaryotic organisms. However, its specific role in controlling the development and virulence of opportunistic pathogenic fungi like A. flavus has remained unclear. Additionally, the underlying mechanism of the TOR pathway remains elusive in the A. flavus. As such, our study provides a useful contribution, as it is the first to comprehensively investigate the majority of genes in the conserved TOR signaling pathway in A. flavus.
Point 2: The authors seemed to perform a series of parallel studies in several genes involved in the TOR pathway in other fungi. However, their data are not properly interconnected to understand the TOR signaling pathway in this fungal pathogen. The authors frequently drew premature conclusions from basic phenotypic observations. For instance, based on their finding that sch9 mutant showed high calcium stress sensitivity, they concluded that Sch9 is the element of the calcineurin-CrzA pathway. Furthermore, based on their finding that the sch9 mutant show weak rapamycin sensitivity and increased Hog1 phosphorylation, they concluded that Sch9 is involved in TOR and HOG pathways. To make such conclusions, the authors should provide more detailed mechanistic data.
Response 2: Yes, we agree with the reviewer's comment. We have carefully reviewed the manuscript and made necessary revisions to eliminate arbitrary conclusions. For example, we have removed the statement that "Sch9 is the element of the calcineurin-CrzA pathway". Furthermore, we have rephrased our conclusions to better reflect our findings. "these results reflected that Sch9 regulates osmotic stress response via the HOG pathway in A. flavus"(Lines 279-280, page 13). We appreciate the reviewer's input, which has contributed to the clarity and accuracy of our work.
Point 3: In the section "Tor kinase plays important roles in A. flavus", some parts of their data are confusing. The authors said they identified a single Tor kinase ortholog, which is orthologous to S. cerevisiae Tor2. And then, they said failed to obtain a null mutant, but constructed a single copy deletion strain delta Tor1+/Tor2-. What does this mean? Does this mean A. flavus diploid strain? So is this heterozygous TOR/tor mutant? Otherwise, does the haploid A. flavus strain they used contain multiple copies of the TOR gene within its genome? What is the real name of A. flavus Tor kinase (Tor1 or Tor2?). "tor1+/tor2-" is the wrong genetic nomenclature. What is the identity of detalTor1+/Tor2-? Please provide detailed information on how all these mutants were generated. A similar issue was found in the analysis of TapA, which is speculated to be essential (what is the deltaTapA1+/TapA2-?). I couldn't find any detailed information even in Materials and Methods. The authors should provide southern blot data to validate all their mutants.
Response 3: Thank you for your comments. We acknowledge the confusion in our presentation and will ensure that accurate genetic nomenclature is used consistently throughout the paper.
In response to your queries, we have included a section in the Materials and Methods, titled "Detection of tor and tapA genes copy number in strains" (Lines 615-621, page 29), to provide details on how we determined the copy numbers of the tor and tapA genes in the strains. Our findings revealed that both the tor and tapA genes are present in double copies in our strains, which guided our decision to construct single-copy deletion strains using homologous recombination. We have verified these copy numbers using absolute quantification PCR (Table S1).
The use of the abbreviation '+/-' for the single copy knockout strains, such as tor+/- and tapA+/-, is consistent with common fungal literature practice. We apologize for any confusion caused by this nomenclature.
Although we did not employ southern blot data for validation, we conducted PCR and gene sequencing to confirm the mutants. We appreciate your comments to improve the clarity and accuracy of our manuscript.
Point 4: How were the FRB domain deletion mutants constructed? If the FKBP12-rapamycin binding (FRB) domain is specifically deleted in the Tor kinase allele, should it be insensitive and resistant to rapamycin? However, the authors showed that the FRB domain deleted TOR allele was indeed non-functional.
Response 4: We appreciate the reviewer's attention to the construction of the Fkbp12-rapamycin binding (FRB) domain deletion mutants and the discrepancy between the expected and observed results.
For the knockout of the FRB domain, we used the homologous recombination method, but because tor genes are double-copy genes, there are also double copies in the FRB domain. Despite our efforts, we encountered challenges in precisely determining the location of the other copy of the tor gene.
We speculate the common expectation that the deletion of the FRB domain should result in insensitivity and resistance to rapamycin, as it disrupts the binding site for Fkbp-rapamycin. However, we observed that the FRB domain-deleted mutant was more sensitive to rapamycin. This intriguing result suggests that there are additional factors or complexities involved in TOR signaling pathway regulation in A. flavus. We hypothesize that this result is related to the double copy of the tor gene. The reviewer's keen observation and comment have contributed to our efforts to better understand and explain this intriguing result.
Point 5: In Figure 4C, the authors should monitor Hog1 phosphorylation patterns under stressed conditions, such as NaCl treatment, and provide quantitative measurements. Similar issues were found in the western blot analysis of Slt2 (Fig. 8D).
Response 5: We agree with the reviewer that we should monitor Hog1 phosphorylation patterns under stressed conditions. In response to this valuable suggestion, we conducted additional experiments to examine Hog1 phosphorylation patterns under NaCl treatment for 30 minutes. The quantitative measurements of Hog1 phosphorylation levels under stress have been added to Figure 4E in the revised manuscript. Similarly, we have addressed the issue raised regarding Slt2 in Figure 8D.
Point 6: For all the deletion mutants generated in this study, the authors should generate complemented strains to validate their data.
Response 6: We appreciate the reviewer's suggestion to generate complemented strains for all the deletion mutants in our study to validate our data. However, due to the extensive number of genes involved in this research, it is hard to create complemented strains for each individual deletion mutant. As suggested by the reviewer, we have constructed complemented strains for several key deletion mutants, such as ΔsitA-C and Δppg1-C.
Response to Reviewer 1 Comments (Recommendations For The Authors):
Point 1: Overall, this manuscript was very poorly organized and not presented logically. It requires extensive English language editing.
Response 1: We appreciate the reviewer's feedback regarding the organization and language quality of our manuscript. To address these concerns, we have restructured the manuscript to improve its logical flow and coherence. We thank the reviewer for their constructive criticism, which has been instrumental in the manuscript's refinement.
Point 2: The authors did not present their figures in the order of description. For example, the authors suddenly described Figure 9A data in lines 128-130 in the middle of describing Figure 1. Furthermore, Figures 1D and 1F were described earlier than Figures 1B and 1C. In addition, Figure S2 was shown earlier than Figure S1. Please check this throughout the manuscript.
Response 2: We thank the reviewer for their insightful observation. We acknowledge the importance of a logical and coherent figure sequence for reader comprehension. After careful review, we have rearranged the text and images throughout the entire document to enhance the reading experience. The revised manuscript now presents figures in a consistent and logical order, following the sequence of descriptions. We believe this improvement will enhance the overall readability and comprehension of our research.
Point 3: The authors should follow the standard genetic nomenclature rules.
Response 3: Thank you for your suggestion. We have revised our manuscript to ensure that we are following the standard genetic nomenclature rules throughout. This includes the correct naming of genes, proteins, and mutations, as well as the use of appropriate italicization and formatting. We follow the rules: gene symbols are typically composed of three lowercase italicized letters, while protein symbols are not italicized, with an initial capital letter followed by lowercase letters.
Point 4: These are just a few examples. Besides the ones that I mentioned, I found numerous grammatically wrong or awkward sentences throughout the manuscript. So this manuscript requires extensive English proofreading.
Response 4: We apologize for the problem of our manuscript. We have asked an English native speaker to enhance the overall language quality and readability of the text. We believe that these improvements will significantly enhance the manuscript's overall quality and make it more accessible to a broader audience.
Response to Reviewer 2 Comments (Public Review):
Point 1: However, findings have not been deeply explored and conclusions mostly are based on parallel phenotypic observations. In addition, there are some concerns that exist surrounding the conclusions.
Response 1: We are grateful for the suggestion. We conduct additional experiments and analyses to delve more deeply into our findings and ensure a more robust basis for our conclusions.
Response to Reviewer 2 Comments (Recommendations For The Authors):
Point 1: Verification for mutants: a single copy deletion strain ΔTor1+/Tor2(containing one copy of the Tor gene), however, in the table of strain list, it seems like null mutants. There are no further verifications for relative genes' expression and no complementary strains.
A. Flavus ΔTor: Δku70; ΔniaD; ΔTor::pyrG
A. Flavus ΔTapA Δku70; ΔniaD; ΔTapA::pyrG
As described in pp208, "While we failed to obtained a null mutant, we constructed a single copy deletion strain ΔTor1+/Tor2- (containing one copy of the Tor gene) constructed by homologous recombination)"? But the authors think there was only one Tor kinase ortholog (AFLA_044350). It is hard to understand for this mutant What is the evidence to verify phenotypes of the ΔTor1+/Tor2- strain resulted from deletion of Tor2, no detail for how to make ΔTor1+/Tor2- strain.
Response 1: Thank you for your important comments and suggestion. We apologize for the confusion caused by genetic nomenclature. We make the necessary corrections in the table of strain lists to accurately reflect the genotypes of the strains (Table S3).
Multicopy variation of genes has not been explored in detail in fungi, especially in A. flavus, but is a commonly known phenomenon in mammalian genomes[1-2]. In yeast, the presence of two tor genes, tor1 and tor2, whereas in higher eukaryotes such as plants, animals, and filamentous fungi, there is only one tor gene[3-4]. The homology comparison results show that the genome of A. flavus contains only one tor gene. However, the tor gene in A. flavus exhibited varying copy numbers, as was confirmed by absolute quantification PCR at the genome level (Table S1).
In this study, we constructed a single copy deletion strain, tor+/-, through homologous recombination. This strain contains one copy of the tor gene. We provide a more detailed and explicit description of the methods used to detect of the genes copy number in strains (Lines 615-621, page 29). We thank the reviewer for pointing out these important issues.
Point 2: For a point mutant strain TORS1904L, they found that the sensitivity to rapamycin is consistent with the WT strain, it could not tell anything. It should be moved to Suppl.
Response 2: Thanks for your important comments. We acknowledge that these results may not provide significant insights. In response to this suggestion, we delete the data related to the TORS1904L point mutant strain and its sensitivity to rapamycin to ensure that the main manuscript focuses on the most pertinent and informative findings. Corresponding modifications have been made in the revised manuscript.
Point 3: For subtitle "Sch9 is correlate with the HOG and TOR pathways "What is the meaning for "correlate" similarly?
Response 3: Thank you for this comment. We apologize for the unclear wording. To enhance clarity, we revise the subtitle to more explicitly convey this conclusion, for example, "The Sch9 kinase is involved in aflatoxin biosynthesis and the HOG pathway". (Lines 242, page 12).
Point 4:for the ΔTapA 1+/TapA 2- strain (containing one copy of the TapA gene). It should have the complementary strain to verify the specific role of TapA. In FigS1B, ΔTOR and ΔTapA it could not tell TOR gene has been edited. Did you test mRNA of TOR gene?
Response 4: Thanks for your important comments. Due to the large number of genes involved, we did not perform a complementation experiment. However, we used PCR and sequencing to verify the editing of our gene. Additionally, we conducted copy number and mRNA analyses to verify its function. The transcriptional level of the tor gene in the tor+/- mutant was downregulated compared to the level in the wild-type strain (Fig. S6).
Response to Reviewer 3 Comments (Public Review):
Point 1: As for many results, I miss the re-complementation of the created mutants throughout the manuscript. This is standard praxis.
Response 1: Thanks for your suggestions. We acknowledge that re-complementation is a standard practice for validating the effects of gene deletions. However, due to the large number of genes involved in our study, we have performed supplementary experiments on a selection of them, such as ΔsitA-C and Δppg1-C. We are grateful to the reviewer for your understanding of this practical consideration.
Point 2: Fig. 1: cultures were grown for 48 h before measuring the transcript level. The authors show that brlA, abaA, and some sexual regulators are less expressed. In my opinion, this does not allow the conclusion that there is a direct control through rapamycin. Since the colonies grow very slowly in the presence of rapamycin, the authors should add rapamycin and follow gene expression after 15, 30, 60, 90 min. The figure legend needs to be more detailed. Which type of cultures were used, liquid, solid medium? Etc.
Response 2: We deeply appreciate the reviewer’s suggestion. Since we found that there were no significant differences in gene expression changes following shorter treatment times, we extended the treatment duration. We conduct additional experiments to examine the gene expression levels at longer time intervals (3, 6, and 9 h) after the addition of rapamycin (Figure 1H-1J). These time points allow us to capture the dynamic changes in gene expression in response to rapamycin more effectively. Additionally, we enhance the figure legend to provide a more comprehensive description that specifies the type of cultures used in the experiments.
Point 3: Why in chapter one Fig. 9 is already cited? Those data should then be included in Fig. 1 for the general phenotype.
Response 3: Thank you for the suggestion. We have reordered the figures in the updated version of the manuscript to ensure that the data for consistent and clarity.
Point 4: The authors wrote that radial growth and conidiation were gradually reduced with increasing rapamycin concentrations. This is not true. There is no gradient! However, it should be tested if there is a gradient if lower concentrations are used. The current data imply that there is a threshold concentration, so either there is 100 % growth or a reduction to 25 %. This looks strange.
Response 4: Thank you for underlining this deficiency. We agree that a threshold concentration versus a gradient is an important distinction that needs to be clarified. Our results show that the addition of excessive quantities of rapamycin does not increase the inhibition of A. flavus growth. As the concentration of the FK506 drug increases, there is a gradual decrease in the growth and cell production of A. flavus. This phenomenon could potentially be attributed to varying mechanisms of action exhibited by the drugs. Therefore, we have revised these confused sentences. ( Lines 120-121, Page 5)
Point 1: There are many wrong spellings:
Fig. 1. Before washed, before washing; RelaTEtive gene expERSion should read relative gene expression. Sclerotial should be sclerotia. See also Fig. 5 F, H, Fig. 6 E. 6D colon diameter should be colony diameter.
Fig. 4E. The expressED level... should read Expression level..... (also without article) Also in A, F, H.
Fig. 6C. TLC detection of WT.... The authors mean AF detection in extracts of WT..... AF was extracted and analyzed by TLC.....
Labelling of axes in one figure should be uniform.
Response 1: Thank you for your reminder. We apologize for the oversights, and we carefully address and correct all the mentioned spelling issues to ensure the accuracy and clarity of the manuscript.
Point 2: If the authors refer to the genes, I think they should be in small letters and italics, if it is the protein, the first letter should be capitalised tap1 (italics) and Tap1.
Response 2: We appreciate this suggestion. We have carefully checked the entire manuscript and revised follow the standard genetic nomenclature rules. We follow the naming conventions for microbial genes and proteins, where gene symbols are typically composed of three lowercase italicized letters, and protein symbols are not italicized, with an initial capital letter followed by lowercase letters.
Point 3: Very often articles are used where I would not use them.
Response 3: Thanks for your careful checks. We are sorry for our carelessness. Based on your comments, we have made the corrections to make the articles harmonized within the whole manuscript. We value the reviewer's feedback, which will contribute to the overall quality of our writing.
References:
[1] Handsaker R, Van Doren, V, Berman, J. et al. Large multiallelic copy number variations in humans. Nat Genet 47, 296–303 (2015).
[2] Wang Y, Wang S, Nie X. et al. Molecular and structural basis of nucleoside diphosphate kinase-mediated regulation of spore and sclerotia development in the fungus Aspergillus flavus. J Biol Chem. 2019 Aug 16;294(33):12415-12431.
[3] Kim DH, Sarbassov DD, Ali SM, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002; 110(2): 163-75.
[4] Fu L, Liu Y, Qin G, et al. The TOR-EIN2 axis mediates nuclear signalling to modulate plant growth. Nature. 2021; 591(7849): 288-292.
-

eLife assessment
This manuscript characterized signaling pathways for growth control and aflatoxin production in the important plant pathogen Aspergillus flavus. Associating tor and tapA with the control of aflatoxin production would be important. However, the copy number of the tor and tapA genes needs to be more clearly established, and without such work, the evidence remains incomplete.
-

Reviewer #1 (Public Review):
Their absolute quantification PCR results with the sumo reference gene led the authors to conclude that A. flavus has two copies of tor and tapA in its genome. However, the the genomic location of the additional copies of tor and tapA are unknown.
I have concerns about the conclusion for the following reasons:
First, the authors should provide more convincing data showing that tor and tapA genes are indeed duplicated genes in A. flavus. The authors appeared to use the A. flavus PTS strain as a parental strain for constructing the tor and tapA mutants. If so, the A. flavus CA14 strain (Hua et al., 2007) should be the parental wild-type strain for the A. flavus PTS strain. I did a BLAST search in NCBI for the torA (AFLA_044350) and tapA (AFLA_092770) genes using the most recent CA14 genome assembly sequence …
Reviewer #1 (Public Review):
Their absolute quantification PCR results with the sumo reference gene led the authors to conclude that A. flavus has two copies of tor and tapA in its genome. However, the the genomic location of the additional copies of tor and tapA are unknown.
I have concerns about the conclusion for the following reasons:
First, the authors should provide more convincing data showing that tor and tapA genes are indeed duplicated genes in A. flavus. The authors appeared to use the A. flavus PTS strain as a parental strain for constructing the tor and tapA mutants. If so, the A. flavus CA14 strain (Hua et al., 2007) should be the parental wild-type strain for the A. flavus PTS strain. I did a BLAST search in NCBI for the torA (AFLA_044350) and tapA (AFLA_092770) genes using the most recent CA14 genome assembly sequence (GCA_014784225.2) and only found one allele for each gene: torA on chromosome 7 and tapA on chromosome 3. I could not find any other parts with similar sequences. Even in another popular A. flavus wild-type strain, NRRL3357, both torA and tapA exist as a single allele. Based on the published genome assembly data for A. flavus, there is no evidence to support the idea that tor and tapA exist as copies of each other. Therefore, the authors could perform a Southern blot analysis to further verify their claim. If torA and tapA indeed exist as duplicate copies in different chromosomal locations, Southern blot data could provide supporting results.
If the tor and tapA genes indeed exist as dual copies, do the duplicate genes have identical DNA and protein sequences? If they have different DNA or protein sequences, they should be named differently as paralogs, such as torA and torB or tapA and tapB.
Second, the authors should consider the possibility of aneuploidy for their constructed mutants. When an essential gene is targeted for deletion, aneuploidy often occurs even in a fungal strain without the "ku" mutation, which results in seemingly dual copies of the gene. As the authors appear to use the A. flavus PTS strain having the "ku" mutation, the parental strain has increased genome instability, which may result in enhanced chromosomal rearrangements. So, it will be necessary to Illumina-sequence their tor and tapA mutants to make sure that they are not aneuploidy.
Furthermore, the genetic nomenclature +/- and -/- should be reserved for heterozygous and homozygous mutants in a diploid strain. As A. flavus is not a diploid strain, this type of description could cause confusion for the readers.
-

Reviewer #2 (Public Review):
In this study, authors identified the complex TOR, HOG and CWI signaling networks-involved genes that relatively modulate the development, aflatoxin biosynthesis and pathogenicity of A. flavus by gene deletions combined with phenotypic observation.
They also analyzed the specific regulatory process and proposed that the TOR signaling pathway interacts with other signaling pathways (MAPK, CWI, calcineurin-CrzA pathway) to regulate the responses to various environmental stresses. Notably, they found that FKBP3 is involved in sclerotia and aflatoxin biosynthesis and rapamycin resistance in A. flavus, especially found that the conserved site K19 of FKBP3 plays a key role in regulating the aflatoxin biosynthesis. In general, there is heavy workload task carried in this study and the findings are interesting and …
Reviewer #2 (Public Review):
In this study, authors identified the complex TOR, HOG and CWI signaling networks-involved genes that relatively modulate the development, aflatoxin biosynthesis and pathogenicity of A. flavus by gene deletions combined with phenotypic observation.
They also analyzed the specific regulatory process and proposed that the TOR signaling pathway interacts with other signaling pathways (MAPK, CWI, calcineurin-CrzA pathway) to regulate the responses to various environmental stresses. Notably, they found that FKBP3 is involved in sclerotia and aflatoxin biosynthesis and rapamycin resistance in A. flavus, especially found that the conserved site K19 of FKBP3 plays a key role in regulating the aflatoxin biosynthesis. In general, there is heavy workload task carried in this study and the findings are interesting and important for understanding or controlling the aflatoxin biosynthesis. However, findings have not been deeply explored and conclusions are mostly are based on parallel phenotypic observations. In addition, there are some concerns for the conclusions.
-

Reviewer #3 (Public Review):
The paper by Li et al. describes the role of the TOR pathway in Aspergillus flavus. The authors tested the effect of rapamycin in WT and different deletion strains. This paper is based on a lot of experiments and work but remains rather descriptive and confirms the results obtained in other fungi. It shows that the TOR pathway is involved in conidiation, aflatoxin production, pathogenicity, and hyphal growth. This is inferred from rapamycin treatment and TOR1/2 deletions. Rapamycin treatment also causes lipid accumulation in hyphae. The phenotypes are not surprising as they have been shown already for several fungi. In addition, one caveat is in my opinion that the strains grow very slowly and this could cause many downstream effects. Several kinases and phosphatases are involved in the TOR pathway. They …
Reviewer #3 (Public Review):
The paper by Li et al. describes the role of the TOR pathway in Aspergillus flavus. The authors tested the effect of rapamycin in WT and different deletion strains. This paper is based on a lot of experiments and work but remains rather descriptive and confirms the results obtained in other fungi. It shows that the TOR pathway is involved in conidiation, aflatoxin production, pathogenicity, and hyphal growth. This is inferred from rapamycin treatment and TOR1/2 deletions. Rapamycin treatment also causes lipid accumulation in hyphae. The phenotypes are not surprising as they have been shown already for several fungi. In addition, one caveat is in my opinion that the strains grow very slowly and this could cause many downstream effects. Several kinases and phosphatases are involved in the TOR pathway. They were known from S. cerevisiae or filamentous fungi. The authors characterized them as well with knock-out approaches.
-

-

eLife assessment
This useful manuscript describes the TOR signaling pathway in the human and plant pathogen Aspergillus flavus. While the authors provide a large amount of descriptive and often confirmative data, the evidence for the new claims made here for the TOR pathway in this species is incomplete.
-

Reviewer #1 (Public Review):
In this manuscript, the authors investigated the roles of the target of rapamycin (TOR) pathway in various pathobiological processes of Aspergillus flavus. They found that rapamycin treatment affects the growth, sporulation, sclerotia, and aflatoxin synthesis of A. flavus. The authors identified four immunophilin genes (FKBP1 -4), among which FKBP3 is involved in both rapamycin and FK506 resistances, with K19 residue being essential for succinylation. The authors identified a single Tor kinase and characterized its function. Subsequently, the authors analyzed a series of downstream effectors of the TOR pathway, including Sch9, TapA, SitA, Ppg1, and Spot7/Nem1, in terms of vegetative growth, sexual development, stress responses, and aflatoxin production.
While the authors provided a large amount of data …
Reviewer #1 (Public Review):
In this manuscript, the authors investigated the roles of the target of rapamycin (TOR) pathway in various pathobiological processes of Aspergillus flavus. They found that rapamycin treatment affects the growth, sporulation, sclerotia, and aflatoxin synthesis of A. flavus. The authors identified four immunophilin genes (FKBP1 -4), among which FKBP3 is involved in both rapamycin and FK506 resistances, with K19 residue being essential for succinylation. The authors identified a single Tor kinase and characterized its function. Subsequently, the authors analyzed a series of downstream effectors of the TOR pathway, including Sch9, TapA, SitA, Ppg1, and Spot7/Nem1, in terms of vegetative growth, sexual development, stress responses, and aflatoxin production.
While the authors provided a large amount of data regarding the genes involved in the TOR pathway, it is highly descriptive and mostly confirmative data, as numerous papers have already shown that the TOR pathway plays essential roles in a myriad of biological processes in multiple fungi. The authors seemed to perform a series of parallel studies in several genes involved in the TOR pathway in other fungi. However, their data are not properly interconnected to understand the TOR signaling pathway in this fungal pathogen. The authors frequently drew premature conclusions from basic phenotypic observations. For instance, based on their finding that sch9 mutant showed high calcium stress sensitivity, they concluded that Sch9 is the element of the calcineurin-CrzA pathway. Furthermore, based on their finding that the sch9 mutant show weak rapamycin sensitivity and increased Hog1 phosphorylation, they concluded that Sch9 is involved in TOR and HOG pathways. To make such conclusions, the authors should provide more detailed mechanistic data.
In the section "Tor kinase plays important roles in A. flavus", some parts of their data are confusing. The authors said they identified a single Tor kinase ortholog, which is orthologous to S. cerevisiae Tor2. And then, they said failed to obtain a null mutant, but constructed a single copy deletion strain delta Tor1+/Tor2-. What does this mean? Does this mean A. flavus diploid strain? So is this heterozygous TOR/tor mutant? Otherwise, does the haploid A. flavus strain they used contain multiple copies of the TOR gene within its genome? What is the real name of A. flavus Tor kinase (Tor1 or Tor2?). "tor1+/tor2-" is the wrong genetic nomenclature. What is the identity of detalTor1+/Tor2-? Please provide detailed information on how all these mutants were generated. A similar issue was found in the analysis of TapA, which is speculated to be essential (what is the deltaTapA1+/TapA2-?). I couldn't find any detailed information even in Materials and Methods. The authors should provide southern blot data to validate all their mutants.
How were the FRB domain deletion mutants constructed? If the FKBP12-rapamycin binding (FRB) domain is specifically deleted in the Tor kinase allele, should it be insensitive and resistant to rapamycin? However, the authors showed that the FRB domain deleted TOR allele was indeed non-functional.
In Figure 4C, the authors should monitor Hog1 phosphorylation patterns under stressed conditions, such as NaCl treatment, and provide quantitative measurements. Similar issues were found in the western blot analysis of Slt2 (Fig. 8D).
For all the deletion mutants generated in this study, the authors should generate complemented strains to validate their data.
-

Reviewer #2 (Public Review):
In this study, the authors identified the complex TOR, HOG, and CWI signaling networks-involved genes that relatively modulate the development, aflatoxin biosynthesis and pathogenicity of A. flavus by gene deletions combined with phenotypic observation.
They also analyzed the specific regulatory process and proposed that the TOR signaling pathway interacts with other signaling pathways (MAPK, CWI, calcineurin-CrzA pathway) to regulate the responses to various environmental stresses. Notably, they found that FKBP3 is involved in sclerotia and aflatoxin biosynthesis and rapamycin resistance in A. flavus, and that the conserved site K19 of FKBP3 plays a key role in regulating the aflatoxin biosynthesis. In general, there is a heavy workload task carried in this study and the findings are interesting and …
Reviewer #2 (Public Review):
In this study, the authors identified the complex TOR, HOG, and CWI signaling networks-involved genes that relatively modulate the development, aflatoxin biosynthesis and pathogenicity of A. flavus by gene deletions combined with phenotypic observation.
They also analyzed the specific regulatory process and proposed that the TOR signaling pathway interacts with other signaling pathways (MAPK, CWI, calcineurin-CrzA pathway) to regulate the responses to various environmental stresses. Notably, they found that FKBP3 is involved in sclerotia and aflatoxin biosynthesis and rapamycin resistance in A. flavus, and that the conserved site K19 of FKBP3 plays a key role in regulating the aflatoxin biosynthesis. In general, there is a heavy workload task carried in this study and the findings are interesting and important for understanding or controlling aflatoxin biosynthesis. However, findings have not been deeply explored and conclusions mostly are based on parallel phenotypic observations. In addition, there are some concerns that exist surrounding the conclusions.
-

Reviewer #3 (Public Review):
The paper by Li et al. describes the role of the TOR pathway in Aspergillus flavus. The authors tested the effect of rapamycin in WT and different deletion strains. This paper is based on a lot of experiments and work but remains rather descriptive and confirms the results obtained in other fungi. It shows that the TOR pathway is involved in conidiation, aflatoxin production, pathogenicity, and hyphal growth. This is inferred from rapamycin treatment and TOR1/2 deletions. Rapamycin treatment also causes lipid accumulation in hyphae. The phenotypes are not surprising as they have been shown already for several fungi. In addition, one caveat is in my opinion that the strains grow very slowly and this could cause many downstream effects. Several kinases and phosphatases are involved in the TOR pathway. They …
Reviewer #3 (Public Review):
The paper by Li et al. describes the role of the TOR pathway in Aspergillus flavus. The authors tested the effect of rapamycin in WT and different deletion strains. This paper is based on a lot of experiments and work but remains rather descriptive and confirms the results obtained in other fungi. It shows that the TOR pathway is involved in conidiation, aflatoxin production, pathogenicity, and hyphal growth. This is inferred from rapamycin treatment and TOR1/2 deletions. Rapamycin treatment also causes lipid accumulation in hyphae. The phenotypes are not surprising as they have been shown already for several fungi. In addition, one caveat is in my opinion that the strains grow very slowly and this could cause many downstream effects. Several kinases and phosphatases are involved in the TOR pathway. They were known from S. cerevisiae or filamentous fungi. The authors characterized them as well with knock-out approaches.
As for many results, I miss the re-complementation of the created mutants throughout the manuscript. This is standard praxis.
Fig. 1: cultures were grown for 48 h before measuring the transcript level. The authors show that brlA, abaA, and some sexual regulators are less expressed. In my opinion, this does not allow the conclusion that there is a direct control through rapamycin. Since the colonies grow very slowly in the presence of rapamycin, the authors should add rapamycin and follow gene expression after 15, 30, 60, 90 min. The figure legend needs to be more detailed. Which type of cultures were used, liquid, solid medium? Etc.
Why in chapter one Fig. 9 is already cited? Those data should then be included in Fig. 1 for the general phenotype.
The authors wrote that radial growth and conidiation were gradually reduced with increasing rapamycin concentrations. This is not true. There is no gradient! However, it should be tested if there is a gradient if lower concentrations are used. The current data imply that there is a threshold concentration, so either there is 100 % growth or a reduction to 25 %. This looks strange.
-