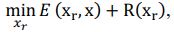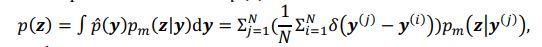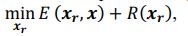Revealing unexpected complex encoding but simple decoding mechanisms in motor cortex via separating behaviorally relevant neural signals
Curation statements for this article:-
Curated by eLife
eLife assessment
This study presents a useful method for the extraction of behaviour-related activity from neural population recordings based on a specific deep learning architecture, a variational autoencoder. Although the authors performed thorough benchmarking of their method in the context of decoding behavioural variables, the evidence supporting claims about encoding is incomplete as the results may stem, in part, from the properties of the method itself.
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
In motor cortex, behaviorally relevant neural responses are entangled with irrelevant signals, which complicates the study of encoding and decoding mechanisms. It remains unclear whether behaviorally irrelevant signals could conceal some critical truth. One solution is to accurately separate behaviorally relevant and irrelevant signals at both single-neuron and single-trial levels, but this approach remains elusive due to the unknown ground truth of behaviorally relevant signals. Therefore, we propose a framework to define, extract, and validate behaviorally relevant signals. Analyzing separated signals in three monkeys performing different reaching tasks, we found neural responses previously considered to contain little information actually encode rich behavioral information in complex nonlinear ways. These responses are critical for neuronal redundancy and reveal movement behaviors occupy a higher-dimensional neural space than previously expected. Surprisingly, when incorporating often-ignored neural dimensions, behaviorally relevant signals can be decoded linearly with comparable performance to nonlinear decoding, suggesting linear readout may be performed in motor cortex. Our findings prompt that separating behaviorally relevant signals may help uncover more hidden cortical mechanisms.
Article activity feed
-

-

-

-

eLife assessment
This study presents a useful method for the extraction of behaviour-related activity from neural population recordings based on a specific deep learning architecture, a variational autoencoder. Although the authors performed thorough benchmarking of their method in the context of decoding behavioural variables, the evidence supporting claims about encoding is incomplete as the results may stem, in part, from the properties of the method itself.
-

Reviewer #1 (Public Review):
This work seeks to understand how behaviour-related information is represented in the neural activity of the primate motor cortex. To this end, a statistical model of neural activity is presented that enables a non-linear separation of behaviour-related from unrelated activity. As a generative model, it enables the separate analysis of these two activity modes, here primarily done by assessing the decoding performance of hand movements the monkeys perform in the experiments. Several lines of analysis are presented to show that while the neurons with significant tuning to movements strongly contribute to the behaviourally-relevant activity subspace, less or un-tuned neurons also carry decodable information. It is further shown that the discovered subspaces enable linear decoding, leading the authors to …
Reviewer #1 (Public Review):
This work seeks to understand how behaviour-related information is represented in the neural activity of the primate motor cortex. To this end, a statistical model of neural activity is presented that enables a non-linear separation of behaviour-related from unrelated activity. As a generative model, it enables the separate analysis of these two activity modes, here primarily done by assessing the decoding performance of hand movements the monkeys perform in the experiments. Several lines of analysis are presented to show that while the neurons with significant tuning to movements strongly contribute to the behaviourally-relevant activity subspace, less or un-tuned neurons also carry decodable information. It is further shown that the discovered subspaces enable linear decoding, leading the authors to conclude that motor cortex read-out can be linear.
Strengths:
In my opinion, using an expressive generative model to analyse neural state spaces is an interesting approach to understand neural population coding. While potentially sacrificing interpretability, this approach allows capturing both redundancies and synergies in the code as done in this paper. The model presented here is a natural non-linear extension of a previous linear model PSID) and uses weak supervision in a manner similar to a previous non-linear model (TNDM).
Weaknesses:
This revised version provides additional evidence to support the author's claims regarding model performance and interpretation of the structure of the resulting latent spaces, in particular the distributed neural code over the whole recorded population, not just the well-tuned neurons. The improved ability to linearly decode behaviour from the relevant subspace and the analysis of the linear subspace projections in my opinion convincingly demonstrates that the model picks up behaviour-relevant dynamics, and that these are distributed widely across the population. As reviewer 3 also points out, I would, however, caution to interpret this as evidence for linear read-out of the motor system - your model performs a non-linear transformation, and while this is indeed linearly decodable, the motor system would need to do something similar first to achieve the same. In fact to me it seems to show the opposite, that behaviour-related information may not be generally accessible to linear decoders (including to down-stream brain areas).
As in my initial review, I would also caution against making strong claims about identifiability although this work and TNDM seem to show that in practise such methods work quite well. CEBRA, in contrast, offers some theoretical guarantees, but it is not a generative model, so would not allow the type of analysis done in this paper. In your model there is a para,eter \alpha to balance between neural and behaviour reconstruction. This seems very similar to TNDM and has to be optimised - if this is correct, then there is manual intervention required to identify a good model.
Somewhat related, I also found that the now comprehensive comparison with related models shows that the using decoding performance (R2) as a metric for model comparison may be problematic: the R2 values reported in Figure 2 (e.g. the MC_RTT dataset) should be compared to the values reported in the neural latent benchmark, which represent well-tuned models (e.g. AutoLFADS). The numbers (difficult to see, a table with numbers in the appendix would be useful, see: https://eval.ai/web/challenges/challenge-page/1256/leaderboard) seem lower than what can be obtained with models without latent space disentanglement. While this does not necessarily invalidate the conclusions drawn here, it shows that decoding performance can depend on a variety of model choices, and may not be ideal to discriminate between models. I'm also surprised by the low neural R2 for LFADS I assume this is condition-averaged) - LFADS tends to perform very well on this metric.
One statement I still cannot follow is how the prior of the variational distribution is modelled. You say you depart from the usual Gaussian prior, but equation 7 seems to suggest there is a normal prior. Are the parameters of this distribution learned? As I pointed out earlier, I however suspect this may not matter much as you give the prior a very low weight. I also still am not sure how you generate a sample from the variational distribution, do you just draw one for each pass?
Summary:
This paper presents a very interesting analysis, but some concerns remain that mainly stem from the complexity of deep learning models. It would be good to acknowledge these as readers without relevant background need to understand where the possible caveats are.
-

Reviewer #2 (Public Review):
Li et al present a method to extract "behaviorally relevant" signals from neural activity. The method is meant to solve a problem which likely has high utility for neuroscience researchers. There are numerous existing methods to achieve this goal some of which the authors compare their method to-thankfully, the revised version includes one of the major previous omissions (TNDM). However, I still believe that d-VAE is a promising approach that has its own advantages. Still, I have issues with the paper as-is. The authors have made relatively few modifications to the text based on my previous comments, and the responses have largely just dismissed my feedback and restated claims from the paper. Nearly all of my previous comments remain relevant for this revised manuscript. As such, they have done little to …
Reviewer #2 (Public Review):
Li et al present a method to extract "behaviorally relevant" signals from neural activity. The method is meant to solve a problem which likely has high utility for neuroscience researchers. There are numerous existing methods to achieve this goal some of which the authors compare their method to-thankfully, the revised version includes one of the major previous omissions (TNDM). However, I still believe that d-VAE is a promising approach that has its own advantages. Still, I have issues with the paper as-is. The authors have made relatively few modifications to the text based on my previous comments, and the responses have largely just dismissed my feedback and restated claims from the paper. Nearly all of my previous comments remain relevant for this revised manuscript. As such, they have done little to assuage my concerns, the most important of which I will restate here using the labels/notation (Q1, Q2, etc) from the reviewer response.
(Q1) I still remain unconvinced that the core findings of the paper are "unexpected". In the response to my previous Specific Comment #1, they say "We use the term 'unexpected' due to the disparity between our findings and the prior understanding concerning neural encoding and decoding." However, they provide no citations or grounding for why they make those claims. What prior understanding makes it unexpected that encoding is more complex than decoding given the entropy, sparseness, and high dimensionality of neural signals (the "encoding") compared to the smoothness and low dimensionality of typical behavioural signals (the "decoding")?
(Q2) I still take issue with the premise that signals in the brain are "irrelevant" simply because they do not correlate with a fixed temporal lag with a particular behavioural feature hand-chosen by the experimenter. In the response to my previous review, the authors say "we employ terms like 'behaviorally-relevant' and 'behaviorally-irrelevant' only regarding behavioral variables of interest measured within a given task, such as arm kinematics during a motor control task.". This is just a restatement of their definition, not a response to my concern, and does not address my concern that the method requires a fixed temporal lag and continual decoding/encoding. My example of reward signals remains. There is a huge body of literature dating back to the 70s on the linear relationships between neural and activity and arm kinematics; in a sense, the authors have chosen the "variable of interest" that proves their point. This all ties back to the previous comment: this is mostly expected, not unexpected, when relating apparently-stochastic, discrete action potential events to smoothly varying limb kinematics.
(Q5) The authors seem to have missed the spirit of my critique: to say "linear readout is performed in motor cortex" is an over-interpretation of what their model can show.
(Q7) Agreeing with my critique is not sufficient; please provide the data or simulations that provides the context for the reference in the fano factor. I believe my critique is still valid.
(Q8) Thank you for comparing to TNDM, it's a useful benchmark.
-

Reviewer #4 (Public Review):
I am a new reviewer for this manuscript, which has been reviewed before. The authors provide a variational autoencoder that has three objectives in the loss: linear reconstruction of behavior from embeddings, reconstruction of neural data, and KL divergence term related to the variational model elements. They take the output of the VAE as the "behaviorally relevant" part of neural data and call the residual "behaviorally irrelevant". Results aim to inspect the linear versus nonlinear behavior decoding using the original raw neural data versus the inferred behaviorally relevant and irrelevant parts of the signal.
Overall, studying neural computations that are behaviorally relevant or not is an important problem, which several previous studies have explored (for example PSID in (Sani et al. 2021), TNDM in …
Reviewer #4 (Public Review):
I am a new reviewer for this manuscript, which has been reviewed before. The authors provide a variational autoencoder that has three objectives in the loss: linear reconstruction of behavior from embeddings, reconstruction of neural data, and KL divergence term related to the variational model elements. They take the output of the VAE as the "behaviorally relevant" part of neural data and call the residual "behaviorally irrelevant". Results aim to inspect the linear versus nonlinear behavior decoding using the original raw neural data versus the inferred behaviorally relevant and irrelevant parts of the signal.
Overall, studying neural computations that are behaviorally relevant or not is an important problem, which several previous studies have explored (for example PSID in (Sani et al. 2021), TNDM in (Hurwitz et al. 2021), TAME-GP in (Balzani et al. 2023), pi-VAE in (Zhou and Wei 2020), and dPCA in (Kobak et al. 2016), etc). However, this manuscript does not properly put their work in the context of such prior works. For example, the abstract states "One solution is to accurately separate behaviorally-relevant and irrelevant signals, but this approach remains elusive", which is not the case given that these prior works have done that. The same is true for various claims in the main text, for example "Furthermore, we found that the dimensionality of primary subspace of raw signals (26, 64, and 45 for datasets A, B, and C) is significantly higher than that of behaviorally-relevant signals (7, 13, and 9), indicating that using raw signals to estimate the neural dimensionality of behaviors leads to an overestimation" (line 321). This finding was presented in (Sani et al. 2021) and (Hurwitz et al. 2021), which is not clarified here. This issue of putting the work in context has been brought up by other reviewers previously but seems to remain largely unaddressed. The introduction is inaccurate also in that it mixes up methods that were designed for separation of behaviorally relevant information with those that are unsupervised and do not aim to do so (e.g., LFADS). The introduction should be significantly revised to explicitly discuss prior models/works that specifically formulated this behavior separation and what these prior studies found, and how this study differs.
Beyond the above, some of the main claims/conclusions made by the manuscript are not properly supported by the analyses and results, which has also been brought up by other reviewers but not fully addressed. First, the analyses here do not support the linear readout from the motor cortex because i) by construction, the VAE here is trained to have a linear readout from its embedding in its loss, which can bias its outputs toward doing well with a linear decoder/readout, and ii) the overall mapping from neural data to behavior includes both the VAE and the linear readout and thus is always nonlinear (even when a linear Kalman filter is used for decoding). This claim is also vague as there is no definition of readout from "motor cortex" or what it means. Why is the readout from the bottleneck of this particular VAE the readout of motor cortex? Second, other claims about properties of individual neurons are also confounded because the VAE is a population-level model that extracts the bottleneck from all neurons. Thus, information can leak from any set of neurons to other sets of neurons during the inference of behaviorally relevant parts of signals. Overall, the results do not convincingly support the claims, and thus the claims should be carefully revised and significantly tempered to avoid misinterpretation by readers.
Below I briefly expand on these as well as other issues, and provide suggestions:
(1) Claims about linearity of "motor cortex" readout are not supported by results yet stated even in the abstract. Instead, what the results support is that for decoding behavior from the output of the dVAE model -- that is trained specifically to have a linear behavior readout from its embedding -- a nonlinear readout does not help. This result can be biased by the very construction of the dVAE's loss that encourages a linear readout/decoding from embeddings and thus does not imply a finding about motor cortex.
(2) Related to the above, it is unclear what the manuscript means by readout from motor cortex. A clearer definition of "readout" (a mapping from what to what?) in general is needed. The mapping that the linearity/nonlinearity claims refer to is from the *inferred* behaviorally relevant neural signals, which themselves are inferred nonlinearly using the VAE. This should be explicitly clarified in all claims, i.e., that only the mapping from distilled signals to behavior is linear, not the whole mapping from neural data to behavior. Again, to say the readout from motor cortex is linear is not supported, including in the abstract.
(3) Claims about individual neurons are also confounded. The d-VAE distilling processing is a population level embedding so the individual distilled neurons are not obtainable on their own without using the population data. This population level approach also raises the possibility that information can leak from one neuron to another during distillation, which is indeed what the authors hope would recover true information about individual neurons that wasn't there in the recording (the pixel denoising example). The authors acknowledge the possibility that information could leak to a neuron that didn't truly have that information and try to rule it out to some extent with some simulations and by comparing the distilled behaviorally relevant signals to the original neural signals. But ultimately, the distilled signals are different enough from the original signals to substantially improve decoding of low information neurons, and one cannot be sure if all of the information in distilled signals from any individual neuron truly belongs to that neuron. It is still quite likely that some of the improved behavior prediction of the distilled version of low-information neurons is due to leakage of behaviorally relevant information from other neurons, not the former's inherent behavioral information. This should be explicitly acknowledged in the manuscript.
(4) Given the nuances involved in appropriate comparisons across methods and since two of the datasets are public, the authors should provide their complete code (not just the dVAE method code), including the code for data loading, data preprocessing, model fitting and model evaluation for all methods and public datasets. This will alleviate concerns and allow readers to confirm conclusions (e.g., figure 2) for themselves down the line.
(5) Related to 1) above, the authors should explore the results if the affine network h(.) (from embedding to behavior) was replaced with a nonlinear ANN. Perhaps linear decoders would no longer be as close to nonlinear decoders. Regardless, the claim of linearity should be revised as described in 1) and 2) above, and all caveats should be discussed.
(6) The beginning of the section on the "smaller R2 neurons" should clearly define what R2 is being discussed. Based on the response to previous reviewers, this R2 "signifies the proportion of neuronal activity variance explained by the linear encoding model, calculated using raw signals". This should be mentioned and made clear in the main text whenever this R2 is referred to.
(7) Various terms require clear definitions. The authors sometimes use vague terminology (e.g., "useless") without a clear definition. Similarly, discussions regarding dimensionality could benefit from more precise definitions. How is neural dimensionality defined? For example, how is "neural dimensionality of specific behaviors" (line 590) defined? Related to this, I agree with Reviewer 2 that a clear definition of irrelevant should be mentioned that clarifies that relevance is roughly taken as "correlated or predictive with a fixed time lag". The analyses do not explore relevance with arbitrary time lags between neural and behavior data.
(8) CEBRA itself doesn't provide a neural reconstruction from its embeddings, but one could obtain one via a regression from extracted CEBRA embeddings to neural data. In addition to decoding results of CEBRA (figure S3), the neural reconstruction of CEBRA should be computed and CEBRA should be added to Figure 2 to see how the behaviorally relevant and irrelevant signals from CEBRA compare to other methods.
References:
Kobak, Dmitry, Wieland Brendel, Christos Constantinidis, Claudia E Feierstein, Adam Kepecs, Zachary F Mainen, Xue-Lian Qi, Ranulfo Romo, Naoshige Uchida, and Christian K Machens. 2016. "Demixed Principal Component Analysis of Neural Population Data." Edited by Mark CW van Rossum. eLife 5 (April): e10989. https://doi.org/10.7554/eLife.10989.
Sani, Omid G., Hamidreza Abbaspourazad, Yan T. Wong, Bijan Pesaran, and Maryam M. Shanechi. 2021. "Modeling Behaviorally Relevant Neural Dynamics Enabled by Preferential Subspace Identification." Nature Neuroscience 24 (1): 140-49. https://doi.org/10.1038/s41593-020-00733-0.
Zhou, Ding, and Xue-Xin Wei. 2020. "Learning Identifiable and Interpretable Latent Models of High-Dimensional Neural Activity Using Pi-VAE." In Advances in Neural Information Processing Systems, 33:7234-47. Curran Associates, Inc. https://proceedings.neurips.cc/paper/2020/hash/510f2318f324cf07fce24c3a4b89c771-Abstract.html.
Hurwitz, Cole, Akash Srivastava, Kai Xu, Justin Jude, Matthew Perich, Lee Miller, and Matthias Hennig. 2021. "Targeted Neural Dynamical Modeling." In Advances in Neural Information Processing Systems. Vol. 34. https://proceedings.neurips.cc/paper/2021/hash/f5cfbc876972bd0d031c8abc37344c28-Abstract.html.
Balzani, Edoardo, Jean-Paul G. Noel, Pedro Herrero-Vidal, Dora E. Angelaki, and Cristina Savin. 2023. "A Probabilistic Framework for Task-Aligned Intra- and Inter-Area Neural Manifold Estimation." In . https://openreview.net/forum?id=kt-dcBQcSA.
-

Author response:
The following is the authors’ response to the previous reviews.
To the Senior Editor and the Reviewing Editor:
We sincerely appreciate the valuable comments provided by the reviewers, the reviewing editor, and the senior editor. Based on our last response and revision, we are confused by the two limitations noted in the eLife assessment.
(1) benchmarking against comparable methods is limited.
In our last revision, we added the comparison experiments with TNDM, as the reviewers requested. Additionally, it is crucial to emphasize that our evaluation of decoding capabilities of behaviorally relevant signals has been benchmarked against the performance of the ANN on raw signals, which, as Reviewer #1 previously noted, nearly represents the upper limit of performance. Consequently, we believe that our benchmarking methods …
Author response:
The following is the authors’ response to the previous reviews.
To the Senior Editor and the Reviewing Editor:
We sincerely appreciate the valuable comments provided by the reviewers, the reviewing editor, and the senior editor. Based on our last response and revision, we are confused by the two limitations noted in the eLife assessment.
(1) benchmarking against comparable methods is limited.
In our last revision, we added the comparison experiments with TNDM, as the reviewers requested. Additionally, it is crucial to emphasize that our evaluation of decoding capabilities of behaviorally relevant signals has been benchmarked against the performance of the ANN on raw signals, which, as Reviewer #1 previously noted, nearly represents the upper limit of performance. Consequently, we believe that our benchmarking methods are sufficiently strong.
(2) some observations may be a byproduct of their method, and may not constitute new scientific observations.
We believe that our experimental results are sufficient to demonstrate that our conclusions are not byproducts of d-VAE based on three reasons:
(1) The d-VAE, as a latent variable model, adheres to the population doctrine, which posits that latent variables are responsible for generating the activities of individual neurons. The goal of such models is to maximize the explanation of the raw signals. At the signal level, the only criterion we can rely on is neural reconstruction performance, in which we have achieved unparalleled results. Thus, it is inappropriate to focus on the mixing process during the model's inference stage while overlooking the crucial de-mixing process during the generation stage and dismissing the significance of our neural reconstruction results. For more details, please refer to the first point in our response to Q4 from Reviewer #4.
(2) The criterion that irrelevant signals should contain minimal information can effectively demonstrate that our conclusions are not by-products of d-VAE. Unfortunately, the reviewers seem to have overlooked this criterion. For more details, please refer to the third point in our response to Q4 from Reviewer #4
(3) Our synthetic experimental results also substantiate that our conclusions are not byproducts of d-VAE. However, it appears the reviewers did not give these results adequate consideration. For more details, please refer to the fourth point in our response to Q4 from Reviewer #4.
Furthermore, our work presents not just "a useful method" but a comprehensive framework. Our study proposes, for the first time, a framework for defining, extracting, and validating behaviorally relevant signals. In our current revision, to clearly distinguish between d-VAE and other methods, we have formalized the extraction of behaviorally relevant signals into a mathematical optimization problem. To our knowledge, current methods have not explicitly proposed extracting behaviorally relevant signals, nor have they identified and addressed the key challenges of extracting relevant signals. Similarly, existing research has not yet defined and validated behaviorally relevant signals. For more details, please refer to our response to Q1 from Reviewer #4.
Based on these considerations, we respectfully request that you reconsider the eLife assessment of our work. We greatly appreciate your time and attention to this matter.
The main revisions made to the manuscript are as follows:
(1) We have formalized the extraction of behaviorally relevant signals into a mathematical optimization problem, enabling a clearer distinction between d-VAE and other models.
(2) We have moderated the assertion about linear readout to highlight its conjectural nature and have broadened the discussion regarding this conclusion.
(3) We have elaborated on the model details of d-VAE and have removed the identifiability claim.
To Reviewer #1
Q1: “As reviewer 3 also points out, I would, however, caution to interpret this as evidence for linear read-out of the motor system - your model performs a non-linear transformation, and while this is indeed linearly decodable, the motor system would need to do something similar first to achieve the same. In fact to me it seems to show the opposite, that behaviour-related information may not be generally accessible to linear decoders (including to down-stream brain areas).”
Thank you for your comments. It's important to note that the conclusions we draw are speculative and not definitive. We use terms like "suggest" to reflect this uncertainty. To further emphasize the conjectural nature of our conclusions, we have deliberately moderated our tone.
The question of whether behaviorally-relevant signals can be accessed by linear decoders or downstream brain regions hinges on the debate over whether the brain employs a strategy of filtering before decoding. If the brain employs such a strategy, the brain can probably access these signals. In our opinion, it is likely that the brain utilizes this strategy.
Given the existence of behaviorally relevant signals, it is reasonable to assume that the brain has intrinsic mechanisms to differentiate between relevant and irrelevant signals. There is growing evidence suggesting that the brain utilizes various mechanisms, such as attention and specialized filtering, to suppress irrelevant signals and enhance relevant signals [1-3]. Therefore, it is plausible that the brain filters before decoding, thereby effectively accessing behaviorally relevant signals.
Thank you for your valuable feedback.
(1) Sreenivasan, Sameet, and Ila Fiete. "Grid cells generate an analog error-correcting code for singularly precise neural computation." Nature neuroscience 14.10 (2011): 1330-1337.
(2) Schneider, David M., Janani Sundararajan, and Richard Mooney. "A cortical filter that learns to suppress the acoustic consequences of movement." Nature 561.7723 (2018): 391-395.
(3) Nakajima, Miho, L. Ian Schmitt, and Michael M. Halassa. "Prefrontal cortex regulates sensory filtering through a basal ganglia-to-thalamus pathway." Neuron 103.3 (2019): 445-458.
Q2: “As in my initial review, I would also caution against making strong claims about identifiability although this work and TNDM seem to show that in practise such methods work quite well. CEBRA, in contrast, offers some theoretical guarantees, but it is not a generative model, so would not allow the type of analysis done in this paper. In your model there is a para,eter \alpha to balance between neural and behaviour reconstruction. This seems very similar to TNDM and has to be optimised - if this is correct, then there is manual intervention required to identify a good model.”
Thank you for your comments.
Considering your concerns about our identifiability claims and the fact that identifiability is not directly relevant to the core of our paper, we have removed content related to identifiability.
Firstly, our model is based on the pi-VAE, which also has theoretical guarantees. However, it is important to note that all such theoretical guarantees (including pi-VAE and CEBRA) are based on certain assumptions that cannot be validated as the true distribution of latent variables remains unknown.
Secondly, it is important to clarify that the identifiability of latent variables does not impact the conclusions of this paper, nor does this paper make specific conclusions about the model's latent variables. Identifiability means that distinct latent variables correspond to distinct observations. If multiple latent variables can generate the same observation, it becomes impossible to determine which one is correct given the observation, which leads to the issue of nonidentifiability. Notably, our analysis focuses on the generated signals, not the latent variables themselves, and thus the identifiability of these variables does not affect our findings.
Our approach, dedicated to extracting these signals, distinctly differs from methods such as TNDM, which focuses on extracting behaviorally relevant latent dynamics. To clearly set apart d-VAE from other models, we have framed the extraction of behaviorally relevant signals as the following mathematical optimization problem:
where 𝑥# denotes generated behaviorally-relevant signals, 𝑥 denotes raw noisy signals, 𝐸(⋅,⋅) demotes reconstruction loss, and 𝑅(⋅) denotes regularization loss. It is important to note that while both d-VAE and TNDM employ reconstruction loss, relying solely on this term is insufficient for determining the optimal degree of similarity between the generated and raw noisy signals. The key to accurately extracting behaviorally relevant signals lies in leveraging prior knowledge about these signals to determine the optimal similarity degree, encapsulated by 𝑅(𝒙𝒓). Other studies have not explicitly proposed extracting behaviorally-relevant signals, nor have they identified and addressed the key challenges involved in extracting relevant signals. Consequently, our approach is distinct from other methods.
Thank you for your valuable feedback.
Q3: “Somewhat related, I also found that the now comprehensive comparison with related models shows that the using decoding performance (R2) as a metric for model comparison may be problematic: the R2 values reported in Figure 2 (e.g. the MC_RTT dataset) should be compared to the values reported in the neural latent benchmark, which represent well-tuned models (e.g. AutoLFADS). The numbers (difficult to see, a table with numbers in the appendix would be useful, see: https://eval.ai/web/challenges/challenge-page/1256/leaderboard) seem lower than what can be obtained with models without latent space disentanglement. While this does not necessarily invalidate the conclusions drawn here, it shows that decoding performance can depend on a variety of model choices, and may not be ideal to discriminate between models. I'm also surprised by the low neural R2 for LFADS I assume this is condition-averaged) - LFADS tends to perform very well on this metric.”
Thank you for your comments. The dataset we utilized is not from the same day as the neural latent benchmark dataset. Notably, there is considerable variation in the length of trials within the RTT paradigm, and the dataset lacks explicit trial information, rendering trial-averaging unsuitable. Furthermore, behaviorally relevant signals are not static averages devoid of variability; even behavioral data exhibits variability. We computed the neural R2 using individual trials rather than condition-averaged responses.
Thank you for your valuable feedback.
Q4: “One statement I still cannot follow is how the prior of the variational distribution is modelled. You say you depart from the usual Gaussian prior, but equation 7 seems to suggest there is a normal prior. Are the parameters of this distribution learned? As I pointed out earlier, I however suspect this may not matter much as you give the prior a very low weight. I also still am not sure how you generate a sample from the variational distribution, do you just draw one for each pass?”
Thank you for your questions.
The conditional distribution of prior latent variables 𝑝%(𝒛|𝒚) is a Gaussian distribution, but the distribution of prior latent variables 𝑝(𝒛) is a mixture Gaussian distribution. The distribution of prior latent variables 𝑝(𝒛) is:
where
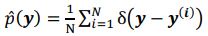 denotes the empirical distribution of behavioral variables
denotes the empirical distribution of behavioral variables𝒚, and 𝑁 denotes the number of samples, 𝒚(𝒊) denotes the 𝒊th sample, δ(⋅) denotes the Dirac delta function, and 𝑝%(𝒛|𝒚) denotes the conditional distribution of prior latent variables given the behavioral variables parameterized by network 𝑚. Based on the above equation, we can see that 𝑝(𝒛) is not a Gaussian distribution, it is a Gaussian mixture model with 𝑁 components, which is theoretically a universal approximator of continuous probability densities.
Learning this prior is important, as illustrated by our latent variable visualizations, which are not a Gaussian distribution. Upon conducting hypothesis testing for both latent variables and behavioral variables, neither conforms to Gaussian distribution (Lilliefors test and Kolmogorov-Smirnov test). Consequently, imposing a constraint on the latent variables towards N(0,1) is expected to affect performance adversely.
Regarding sampling, during training process, we draw only one sample from the approximate posterior distribution
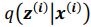 . It is worth noting that drawing multiple samples or one sample for each pass does not affect the experimental results. After training, we can generate a sample from the prior by providing input behavioral data 𝒚(𝒊) and then generating corresponding samples via
. It is worth noting that drawing multiple samples or one sample for each pass does not affect the experimental results. After training, we can generate a sample from the prior by providing input behavioral data 𝒚(𝒊) and then generating corresponding samples via 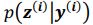 and
and 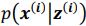 . To extract behaviorally-relevant signals from raw signals, we use
. To extract behaviorally-relevant signals from raw signals, we use 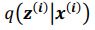 and
and 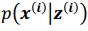 .
.Thank you for your valuable feedback.
Q5: “(1) I found the figures good and useful, but the text is, in places, not easy to follow. I think the manuscript could be shortened somewhat, and in some places more concise focussed explanations would improve readability.
(2) I would not call the encoding "complex non-linear" - non-linear is a clear term, but complex can mean many things (e.g. is a quadratic function complex?) ”
Thank you for your recommendation. We have revised the manuscript for enhanced clarity. We call the encoding “complex nonlinear” because neurons encode information with varying degrees of nonlinearity, as illustrated in Fig. 3b, f, and Fig. S3b.
Thank you for your valuable feedback.
To Reviewer #2
Q1: “I still remain unconvinced that the core findings of the paper are "unexpected". In the response to my previous Specific Comment #1, they say "We use the term 'unexpected' due to the disparity between our findings and the prior understanding concerning neural encoding and decoding." However, they provide no citations or grounding for why they make those claims. What prior understanding makes it unexpected that encoding is more complex than decoding given the entropy, sparseness, and high dimensionality of neural signals (the "encoding") compared to the smoothness and low dimensionality of typical behavioural signals (the "decoding")?”
Thank you for your comments. We believe that both the complexity of neural encoding and the simplicity of neural decoding in motor cortex are unexpected.
The Complexity of Neural Encoding: As noted in the Introduction, neurons with small R2 values were traditionally considered noise and consequently disregarded, as detailed in references [1-3]. However, after filtering out irrelevant signals, we discovered that these neurons actually contain substantial amounts of behavioral information, previously unrecognized. Similarly, in population-level analyses, neural signals composed of small principal components (PCs) are often dismissed as noise, with analyses typically utilizing only between 6 and 18 PCs [4-10]. Yet, the discarded PC signals nonlinearly encode significant amounts of information, with practically useful dimensions found to range between 30 and 40—far exceeding the usual number analyzed. These findings underscore the complexity of neural encoding and are unexpected.
The Simplicity of Neural Decoding: In the motor cortex, nonlinear decoding of raw signals has been shown to significantly outperform linear decoding, as evidenced in references [11,12]. Interestingly, after separating behaviorally relevant and irrelevant signals, we observed that the linear decoding performance of behaviorally relevant signals is nearly equivalent to that of nonlinear decoding—a phenomenon previously undocumented in the motor cortex. This discovery is also unexpected.
Thank you for your valuable feedback.
(1) Georgopoulos, Apostolos P., Andrew B. Schwartz, and Ronald E. Kettner. "Neuronal population coding of movement direction." Science 233.4771 (1986): 1416-1419.
(2) Hochberg, Leigh R., et al. "Reach and grasp by people with tetraplegia using a neurally controlled robotic arm." Nature 485.7398 (2012): 372-375.
(3) Inoue, Yoh, et al. "Decoding arm speed during reaching." Nature communications 9.1 (2018): 5243.
(4) Churchland, Mark M., et al. "Neural population dynamics during reaching." Nature 487.7405 (2012): 51-56.
(5) Kaufman, Matthew T., et al. "Cortical activity in the null space: permitting preparation without movement." Nature neuroscience 17.3 (2014): 440-448.
(6) Elsayed, Gamaleldin F., et al. "Reorganization between preparatory and movement population responses in motor cortex." Nature communications 7.1 (2016): 13239.
(7) Sadtler, Patrick T., et al. "Neural constraints on learning." Nature 512.7515 (2014): 423426.
(8) Golub, Matthew D., et al. "Learning by neural reassociation." Nature neuroscience 21.4 (2018): 607-616.
(9) Gallego, Juan A., et al. "Cortical population activity within a preserved neural manifold underlies multiple motor behaviors." Nature communications 9.1 (2018): 4233.
(10) Gallego, Juan A., et al. "Long-term stability of cortical population dynamics underlying consistent behavior." Nature neuroscience 23.2 (2020): 260-270.
(11) Glaser, Joshua I., et al. "Machine learning for neural decoding." Eneuro 7.4 (2020).
(12) Willsey, Matthew S., et al. "Real-time brain-machine interface in non-human primates achieves high-velocity prosthetic finger movements using a shallow feedforward neural network decoder." Nature Communications 13.1 (2022): 6899.
Q2: “I still take issue with the premise that signals in the brain are "irrelevant" simply because they do not correlate with a fixed temporal lag with a particular behavioural feature handchosen by the experimenter. In the response to my previous review, the authors say "we employ terms like 'behaviorally-relevant' and 'behaviorally-irrelevant' only regarding behavioral variables of interest measured within a given task, such as arm kinematics during a motor control task.". This is just a restatement of their definition, not a response to my concern, and does not address my concern that the method requires a fixed temporal lag and continual decoding/encoding. My example of reward signals remains. There is a huge body of literature dating back to the 70s on the linear relationships between neural and activity and arm kinematics; in a sense, the authors have chosen the "variable of interest" that proves their point. This all ties back to the previous comment: this is mostly expected, not unexpected, when relating apparently-stochastic, discrete action potential events to smoothly varying limb kinematics.”
Thank you for your comments.
Regarding the experimenter's specification of behavioral variables of interest, we followed common practice in existing studies [1, 2]. Regarding the use of fixed temporal lags, we followed the same practice as papers related to the dataset we use, which assume fixed temporal lags [3-5]. Furthermore, many studies in the motor cortex similarly use fixed temporal lags [68].
Concerning the issue of rewards, in the paper you mentioned [9], the impact of rewards occurs after the reaching phase. It's important to note that in our experiments, we analyze only the reaching phase, without any post-movement phase.
If the impact of rewards can be stably reflected in the signals in the reaching phase of the subsequent trial, and if the reward-induced signals do not interfere with decoding—since these signals are harmless for decoding and beneficial for reconstruction—our model is likely to capture these signals. If the signals induced by rewards during the reaching phase are randomly unstable, our model will likely be unable to capture them.
If the goal is to extract post-movement neural activity from both rewarded and unrewarded trials, and if the neural patterns differ between these conditions, one could replace the d-VAE's regression loss, used for continuous kinematics decoding, with a classification loss tailored to distinguish between rewarded and unrewarded conditions.
To clarify the definition, we have revised it in the manuscript. Specifically, before a specific definition, we briefly introduce the relevant signals and irrelevant signals. Behaviorally irrelevant signals refer to those not directly associated with the behavioral variables of interest and may include noise or signals from variables of no interest. In contrast, behaviorally relevant signals refer to those directly related to the behavioral variables of interest. For instance, rewards in the post-movement phase are not directly related to behavioral variables (kinematics) in the reaching movement phase.
It is important to note that our definition of behaviorally relevant signals not only includes decoding capabilities but also specific requirement at the signal level, based on two key requirements:
(1) they should closely resemble raw signals to preserve the underlying neuronal properties without becoming so similar that they include irrelevant signals. (encoding requirement), and (2) they should contain behavioral information as much as possible (decoding requirement). Signals that meet both requirements are considered effective behaviorally relevant signals. In our study, we assume raw signals are additively composed of behaviorally-relevant and irrelevant signals. We define irrelevant signals as those remaining after subtracting relevant signals from raw signals. Therefore, we believe our definition is clearly articulated.
Thank you for your valuable feedback.
(1) Sani, Omid G., et al. "Modeling behaviorally relevant neural dynamics enabled by preferential subspace identification." Nature Neuroscience 24.1 (2021): 140-149.
(2) Buetfering, Christina, et al. "Behaviorally relevant decision coding in primary somatosensory cortex neurons." Nature neuroscience 25.9 (2022): 1225-1236.
(3) Wang, Fang, et al. "Quantized attention-gated kernel reinforcement learning for brain– machine interface decoding." IEEE transactions on neural networks and learning systems 28.4 (2015): 873-886.
(4) Dyer, Eva L., et al. "A cryptography-based approach for movement decoding." Nature biomedical engineering 1.12 (2017): 967-976.
(5) Ahmadi, Nur, Timothy G. Constandinou, and Christos-Savvas Bouganis. "Robust and accurate decoding of hand kinematics from entire spiking activity using deep learning." Journal of Neural Engineering 18.2 (2021): 026011.
(6) Churchland, Mark M., et al. "Neural population dynamics during reaching." Nature 487.7405 (2012): 51-56.
(7) Kaufman, Matthew T., et al. "Cortical activity in the null space: permitting preparation without movement." Nature neuroscience 17.3 (2014): 440-448.
(8) Elsayed, Gamaleldin F., et al. "Reorganization between preparatory and movement population responses in motor cortex." Nature communications 7.1 (2016): 13239.
(9) Ramkumar, Pavan, et al. "Premotor and motor cortices encode reward." PloS one 11.8 (2016): e0160851.
Q3: “The authors seem to have missed the spirit of my critique: to say "linear readout is performed in motor cortex" is an over-interpretation of what their model can show.”
Thank you for your comments. It's important to note that the conclusions we draw are speculative and not definitive. We use terms like "suggest" to reflect this uncertainty. To further emphasize the conjectural nature of our conclusions, we have deliberately moderated our tone.
The question of whether behaviorally-relevant signals can be accessed by downstream brain regions hinges on the debate over whether the brain employs a strategy of filtering before decoding. If the brain employs such a strategy, the brain can probably access these signals. In our view, it is likely that the brain utilizes this strategy.
Given the existence of behaviorally relevant signals, it is reasonable to assume that the brain has intrinsic mechanisms to differentiate between relevant and irrelevant signals. There is growing evidence suggesting that the brain utilizes various mechanisms, such as attention and specialized filtering, to suppress irrelevant signals and enhance relevant signals [1-3]. Therefore, it is plausible that the brain filters before decoding, thereby effectively accessing behaviorally relevant signals.
Regarding the question of whether the brain employs linear readout, given the limitations of current observational methods and our incomplete understanding of brain mechanisms, it is challenging to ascertain whether the brain employs a linear readout. In many cortical areas, linear decoders have proven to be sufficiently accurate. Consequently, numerous studies [4, 5, 6], including the one you referenced [4], directly employ linear decoders to extract information and formulate conclusions based on the decoding results. Contrary to these approaches, our research has compared the performance of linear and nonlinear decoders on behaviorally relevant signals and found their decoding performance is comparable. Considering both the decoding accuracy and model complexity, our results suggest that the motor cortex may utilize linear readout to decode information from relevant signals. Given the current technological limitations, we consider it reasonable to analyze collected data to speculate on the potential workings of the brain, an approach that many studies have also embraced [7-10]. For instance, a study [7] deduces strategies the brain might employ to overcome noise by analyzing the structure of recorded data and decoding outcomes for new stimuli.
Thank you for your valuable feedback.
(1) Sreenivasan, Sameet, and Ila Fiete. "Grid cells generate an analog error-correcting code for singularly precise neural computation." Nature neuroscience 14.10 (2011): 1330-1337.
(2) Schneider, David M., Janani Sundararajan, and Richard Mooney. "A cortical filter that learns to suppress the acoustic consequences of movement." Nature 561.7723 (2018): 391-395.
(3) Nakajima, Miho, L. Ian Schmitt, and Michael M. Halassa. "Prefrontal cortex regulates sensory filtering through a basal ganglia-to-thalamus pathway." Neuron 103.3 (2019): 445-458.
(4) Jurewicz, Katarzyna, et al. "Irrational choices via a curvilinear representational geometry for value." bioRxiv (2022): 2022-03.
(5) Hong, Ha, et al. "Explicit information for category-orthogonal object properties increases along the ventral stream." Nature neuroscience 19.4 (2016): 613-622.
(6) Chang, Le, and Doris Y. Tsao. "The code for facial identity in the primate brain." Cell 169.6 (2017): 1013-1028.
(7) Ganmor, Elad, Ronen Segev, and Elad Schneidman. "A thesaurus for a neural population code." Elife 4 (2015): e06134.
(8) Churchland, Mark M., et al. "Neural population dynamics during reaching." Nature 487.7405 (2012): 51-56.
(9) Gallego, Juan A., et al. "Cortical population activity within a preserved neural manifold underlies multiple motor behaviors." Nature communications 9.1 (2018): 4233.
(10) Gallego, Juan A., et al. "Long-term stability of cortical population dynamics underlying consistent behavior." Nature neuroscience 23.2 (2020): 260-270.
Q4: “Agreeing with my critique is not sufficient; please provide the data or simulations that provides the context for the reference in the fano factor. I believe my critique is still valid.”
Thank you for your comments. As we previously replied, Churchland's research examines the variability of neural signals across different stages, including the preparation and execution phases, as well as before and after the target appears. Our study, however, focuses exclusively on the movement execution phase. Consequently, we are unable to produce comparative displays similar to those in his research. Intuitively, one might expect that the variability of behaviorally relevant signals would be lower; however, since no prior studies have accurately extracted such signals, the specific FF values of behaviorally relevant signals remain unknown. Therefore, presenting these values is meaningful, and can provide a reference for future research. While we cannot compare FF across different stages, we can numerically compare the values to the Poisson count process. An FF of 1 indicates a Poisson firing process, and our experimental data reveals that most neurons have an FF less than 1, indicating that the variance in firing counts is below the mean. Thank you for your valuable feedback.
To Reviewer #4
Q1: “Overall, studying neural computations that are behaviorally relevant or not is an important problem, which several previous studies have explored (for example PSID in (Sani et al. 2021), TNDM in (Hurwitz et al. 2021), TAME-GP in (Balzani et al. 2023), pi-VAE in (Zhou and Wei 2020), and dPCA in (Kobak et al. 2016), etc). However, this manuscript does not properly put their work in the context of such prior works. For example, the abstract states "One solution is to accurately separate behaviorally-relevant and irrelevant signals, but this approach remains elusive", which is not the case given that these prior works have done that. The same is true for various claims in the main text, for example "Furthermore, we found that the dimensionality of primary subspace of raw signals (26, 64, and 45 for datasets A, B, and C) is significantly higher than that of behaviorally-relevant signals (7, 13, and 9), indicating that using raw signals to estimate the neural dimensionality of behaviors leads to an overestimation" (line 321). This finding was presented in (Sani et al. 2021) and (Hurwitz et al. 2021), which is not clarified here. This issue of putting the work in context has been brought up by other reviewers previously but seems to remain largely unaddressed. The introduction is inaccurate also in that it mixes up methods that were designed for separation of behaviorally relevant information with those that are unsupervised and do not aim to do so (e.g., LFADS). The introduction should be significantly revised to explicitly discuss prior models/works that specifically formulated this behavior separation and what these prior studies found, and how this study differs.”
Thank you for your comments. Our statement about “One solution is to accurately separate behaviorally-relevant and irrelevant signals, but this approach remains elusive” is accurate. To our best knowledge, there is no prior works to do this work--- separating accurate behaviorally relevant neural signals at both single-neuron and single-trial resolution. The works you mentioned have not explicitly proposed extracting behaviorally relevant signals, nor have they identified and addressed the key challenges of extracting relevant signals, namely determining the optimal degree of similarity between the generated relevant signals and raw signals. Those works focus on the latent neural dynamics, rather than signal level.
To clearly set apart d-VAE from other models, we have framed the extraction of behaviorally relevant signals as the following mathematical optimization problem:
where 𝒙𝒓 denotes generated behaviorally-relevant signals, 𝒙 denotes raw noisy signals, 𝐸(⋅,⋅) demotes reconstruction loss, and 𝑅(⋅) denotes regularization loss. It is important to note that while both d-VAE and TNDM employ reconstruction loss, relying solely on this term is insufficient for determining the optimal degree of similarity between the generated and raw noisy signals. The key to accurately extracting behaviorally relevant signals lies in leveraging prior knowledge about these signals to determine the optimal similarity degree, encapsulated by 𝑅(𝒙𝒓). All the works you mentioned did not have the key part 𝑅(𝒙𝒓).
Regarding the dimensionality estimation, the dimensionality of neural manifolds quantifies the degrees of freedom required to describe population activity without significant information loss.
There are two differences between our work and PSID and TNDM.
First, the dimensions they refer to are fundamentally different from ours. The dimensionality we describe pertains to a linear subspace, where a neural dimension or neural mode or principal component basis,
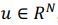 , with N representing the number of neurons. However, the vector length of a neural mode of PSID and our approach differs; PSID requires concatenating multiple time steps T, essentially making
, with N representing the number of neurons. However, the vector length of a neural mode of PSID and our approach differs; PSID requires concatenating multiple time steps T, essentially making 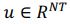 , TNDM, on the other hand, involves nonlinear dimensionality reduction, which is different from linear dimensionality reduction.
, TNDM, on the other hand, involves nonlinear dimensionality reduction, which is different from linear dimensionality reduction.Second, we estimate neural dimensionality by explaining the variance of neural signals, whereas PSID and TNDM determine dimensionality through decoding performance saturation. It is important to note that the dimensionality at which decoding performance saturates may not accurately reflect the true dimensionality of neural manifolds, as some dimensions may contain redundant information that does not enhance decoding performance.
We acknowledge that while LFADS can generate signals that contain some behavioral information, it was not specifically designed to do so. Following your suggestion, we have removed this reference from the Introduction.
Thank you for your valuable feedback.
Q2: “Claims about linearity of "motor cortex" readout are not supported by results yet stated even in the abstract. Instead, what the results support is that for decoding behavior from the output of the dVAE model -- that is trained specifically to have a linear behavior readout from its embedding -- a nonlinear readout does not help. This result can be biased by the very construction of the dVAE's loss that encourages a linear readout/decoding from embeddings, and thus does not imply a finding about motor cortex.”
Thank you for your comments. We respectfully disagree with the notion that the ability of relevant signals to be linearly decoded is due to constraints that allow embedding to be linearly decoded. Embedding involves reorganizing or transforming the structure of original signals, and they can be linearly decoded does not mean the corresponding signals can be decoded linearly.
Let's clarify this with three intuitive examples:
Example 1: Image denoising is a well-established field. Whether employing supervised or blind denoising methods [1, 2], both can effectively recover the original image. This denoising process closely resembles the extraction of behaviorally relevant signals from raw signals. Consider if noisy images are not amenable to linear decoding (classification); would removing the noise enable linear decoding? The answer is no. Typically, the noise in images captured under normal conditions is minimal, yet even the clear images remain challenging to decode linearly.
Example 2: Consider the task of face recognition, where face images are set against various backgrounds, in this context, the pixels representing the face corresponds to relevant signals, while the background pixels are considered irrelevant. Suppose a network is capable of extracting the face pixels and the resulting embedding can be linearly decoded. Can the face pixels themselves be linearly decoded? The answer is no. If linear decoding of face pixels were feasible, the challenging task of face recognition could be easily resolved by merely extracting the face from the background and training a linear classifier.
Example 3: In the MNIST dataset, the background is uniformly black, and its impact is minimal. However, linear SVM classifiers used directly on the original pixels significantly underperform compared to non-linear SVMs.
In summary, embedding involves reorganizing the structure of the original signals through a feature transformation function. However, the reconstruction process can recover the structure of the original signals from the embedding. The fact that the structure of the embedding can be linearly decoded does not imply that the structure of the original signals can be linearly decoded in the same way. It is inappropriate to focus on the compression process without equally considering the reconstruction process.
Thank you for your valuable feedback.
(1) Mao, Xiao-Jiao, Chunhua Shen, and Yu-Bin Yang. "Image restoration using convolutional auto-encoders with symmetric skip connections." arXiv preprint arXiv:1606.08921 (2016).
(2) Lehtinen, Jaakko, et al. "Noise2Noise: Learning image restoration without clean data." International Conference on Machine Learning. International Machine Learning Society, 2018.
Q3: “Related to the above, it is unclear what the manuscript means by readout from motor cortex. A clearer definition of "readout" (a mapping from what to what?) in general is needed. The mapping that the linearity/nonlinearity claims refer to is from the *inferred* behaviorally relevant neural signals, which themselves are inferred nonlinearly using the VAE. This should be explicitly clarified in all claims, i.e., that only the mapping from distilled signals to behavior is linear, not the whole mapping from neural data to behavior. Again, to say the readout from motor cortex is linear is not supported, including in the abstract.”
Thank you for your comments. We have revised the manuscript to make it more clearly. Thank you for your valuable feedback.
Q4: “Claims about individual neurons are also confounded. The d-VAE distilling processing is a population level embedding so the individual distilled neurons are not obtainable on their own without using the population data. This population level approach also raises the possibility that information can leak from one neuron to another during distillation, which is indeed what the authors hope would recover true information about individual neurons that wasn't there in the recording (the pixel denoising example). The authors acknowledge the possibility that information could leak to a neuron that didn't truly have that information and try to rule it out to some extent with some simulations and by comparing the distilled behaviorally relevant signals to the original neural signals. But ultimately, the distilled signals are different enough from the original signals to substantially improve decoding of low information neurons, and one cannot be sure if all of the information in distilled signals from any individual neuron truly belongs to that neuron. It is still quite likely that some of the improved behavior prediction of the distilled version of low-information neurons is due to leakage of behaviorally relevant information from other neurons, not the former's inherent behavioral information. This should be explicitly acknowledged in the manuscript.”
Thank you for your comments. We value your insights regarding the mixing process. However, we are confident in the robustness of our conclusions. We respectfully disagree with the notion that the small R2 values containing significant information are primarily due to leakage, and we base our disagreement on four key reasons.
(1) Neural reconstruction performance is a reliable and valid criterion.
The purpose of latent variable models is to explain neuronal activity as much as possible. Given the fact that the ground truth of behaviorally-relevant signals, the latent variables, and the generative model is unknow, it becomes evident that the only reliable reference at the signal level is the raw signals. A crucial criterion for evaluating the reliability of latent variable models (including latent variables and generated relevant signals) is their capability to effectively explain the raw signals [1]. Consequently, we firmly maintain the belief that if the generated signals closely resemble the raw signals to the greatest extent possible, in accordance with an equivalence principle, we can claim that these obtained signals faithfully retain the inherent properties of single neurons.
Reviewer #4 appears to focus on the compression (mixing) process without giving equal consideration to the reconstruction (de-mixing) process. Numerous studies have demonstrated that deep autoencoders can reconstruct the original signal very effectively. For example, in the field of image denoising, autoencoders are capable of accurately restoring the original image [2, 3]. If one persistently focuses on the fact of mixing and ignores the reconstruction (demix) process, even if the only criterion that we can rely on at the signal level is high, one still won't acknowledge it. If this were the case, many problems would become unsolvable. For instance, a fundamental criterion for latent variable models is their ability to explain the original data. If the ground truth of the latent variables remains unknown and the reconstruction criterion is disregarded, how can we validate the effectiveness of the model, the validity of the latent variables, or ensure that findings related to latent variables are not merely by-products of the model? Therefore, we disagree with the aforementioned notion. We believe that as long as the reconstruction performance is satisfactory, the extracted signals have successfully retained the characteristics of individual neurons.
In our paper, we have shown in various ways that our generated signals sufficiently resemble the raw signals, including visualizing neuronal activity (Fig. 2m, Fig. 3i, and Fig. S5), achieving the highest performance among competitors (Fig. 2d, h, l), and conducting control analyses. Therefore, we believe our results are reliable.
(1) Cunningham, J.P. and Yu, B.M., 2014. Dimensionality reduction for large-scale neural recordings. Nature neuroscience, 17(11), pp.1500-1509.
(2) Mao, Xiao-Jiao, Chunhua Shen, and Yu-Bin Yang. "Image restoration using convolutional auto-encoders with symmetric skip connections." arXiv preprint arXiv:1606.08921 (2016).
(3) Lehtinen, Jaakko, et al. "Noise2Noise: Learning image restoration without clean data." International Conference on Machine Learning. International Machine Learning Society, 2018.
(2) There is no reason for d-VAE to add signals that do not exist in the original signals.
(1) Adding signals that does not exist in the small R2 neurons would decrease the reconstruction performance. This is because if the added signals contain significant information, they will not resemble the irrelevant signals which contain no information, and thus, the generated signals will not resemble the raw signals. The model optimizes towards reducing the reconstruction loss, and this scenario deviates from the model's optimization direction. It is worth mentioning that when the model only has reconstruction loss without the interference of decoding loss, we believe that information leakage does not happen. Because the model can only be optimized in a direction that is similar to the raw signals; adding non-existent signals to the generated signals would increase the reconstruction loss, which is contrary to the objective of optimization.
(2) Information carried by these additional signals is redundant for larger R2 neurons, thus they do not introduce new information that can enhance the decoding performance of the neural population, which does not benefit the decoding loss.
Based on these two points, we believe the model would not perform such counterproductive and harmful operations.
(3) The criterion that irrelevant signals should contain minimal information can effectively rule out the leakage scenario.
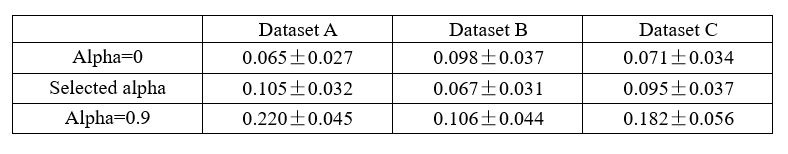
The criterion that irrelevant signals should contain minimal information is very important, but it seems that reviewer #4 has continuously overlooked their significance. If the model's reconstruction is insufficient, or if additional information is added (which we do not believe will happen), the residuals would decode a large amount of information, and this criterion would exclude selecting such signals. To clarify, if we assume that x, y, and z denote the raw, relevant, and irrelevant signals of smaller R2 neurons, with x=y+z, and the extracted relevant signals become y+m, the irrelevant signals become z-m in this case. Consequently, the irrelevant signals contain a significant amount of information.
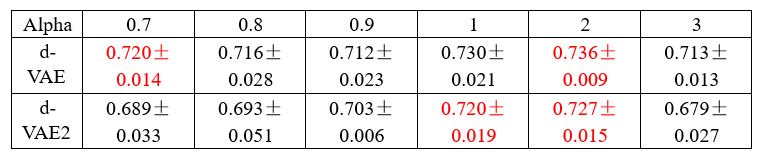
We presented the decoding R2 for irrelevant signals in real datasets under three distillation scenarios: a bias towards reconstruction (alpha=0, an extreme case where the model only has reconstruction loss without decoding loss), a balanced trade-off, and a bias towards decoding (alpha=0.9), as detailed in Table 1. If significant information from small R2 neurons leaks from large R2 neurons, the irrelevant signals should contain a large amount of information. However, our results indicate that the irrelevant signals contain only minimal information, and their performance closely resembles that of the model training solely with reconstruction loss, showing no significant differences (P > 0.05, Wilcoxon rank-sum test). When the model leans towards decoding, some useful information will be left in the residuals, and irrelevant signals will contain a substantial amount of information, as observed in Table 1, alpha=0.9. Therefore, we will not choose these signals for analysis.
In conclusion, the criterion that irrelevant signals should contain minimal information is a very effective measure to exclude undesirable signals.
Author response table 1.
Decoding R2 of irrelevant signals
(4) Synthetic experiments can effectively rule out the leakage scenario.
In the absence of ground truth data, synthetic experiments serve as an effective method for validating models and are commonly employed [1-3].
Our experimental results demonstrate that d-VAE can effectively extract neural signals that more closely resemble actual behaviorally relevant signals (Fig. S2g). If there were information leakage, it would decrease the similarity to the ground truth signals, hence we have ruled out this possibility. Moreover, in synthetic experiments with small R2 neurons (Fig. S10), results also demonstrate that our model could make these neurons more closely resemble ground truth relevant signals and recover their information.
In summary, synthetic experiments strongly demonstrate that our model can recover obscured neuronal information, rather than adding signals that do not exist.
(1) Pnevmatikakis, Eftychios A., et al. "Simultaneous denoising, deconvolution, and demixing of calcium imaging data." Neuron 89.2 (2016): 285-299.
(2) Schneider, Steffen, Jin Hwa Lee, and Mackenzie Weygandt Mathis. "Learnable latent embeddings for joint behavioural and neural analysis." Nature 617.7960 (2023): 360-368.
(3) Zhou, Ding, and Xue-Xin Wei. "Learning identifiable and interpretable latent models of high-dimensional neural activity using pi-VAE." Advances in Neural Information Processing Systems 33 (2020): 7234-7247.
Based on these four points, we are confident in the reliability of our results. If Reviewer #4 considers these points insufficient, we would highly appreciate it if specific concerns regarding any of these aspects could be detailed.
Thank you for your valuable feedback.
Q5: “Given the nuances involved in appropriate comparisons across methods and since two of the datasets are public, the authors should provide their complete code (not just the dVAE method code), including the code for data loading, data preprocessing, model fitting and model evaluation for all methods and public datasets. This will alleviate concerns and allow readers to confirm conclusions (e.g., figure 2) for themselves down the line.”
Thanks for your suggestion.
Our codes are now available on GitHub at https://github.com/eric0li/d-VAE. Thank you for your valuable feedback.
Q6: “Related to 1) above, the authors should explore the results if the affine network h(.) (from embedding to behavior) was replaced with a nonlinear ANN. Perhaps linear decoders would no longer be as close to nonlinear decoders. Regardless, the claim of linearity should be revised as described in 1) and 2) above, and all caveats should be discussed.”
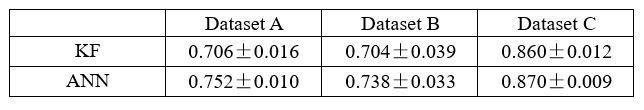
Thank you for your suggestion. We appreciate your feasible proposal that can be empirically tested. Following your suggestion, we have replaced the decoding of the latent variable z to behavior y with a nonlinear neural network, specifically a neural network with a single hidden layer. The modified model is termed d-VAE2. We applied the d-VAE2 to the real data, and selected the optimal alpha through the validation set. As shown in Table 1, results demonstrate that the performance of KF and ANN remains comparable. Therefore, the capacity to linearly decode behaviorally relevant signals does not stem from the linear decoding of embeddings.
Author response table 2.
Decoding R2 of behaviorally relevant signals obtained by d-VAE2
Additionally, it is worth noting that this approach is uncommon and is considered somewhat inappropriate according to the Information Bottleneck theory [1]. According to the Information Bottleneck theory, information is progressively compressed in multilayer neural networks, discarding what is irrelevant to the output and retaining what is relevant. This means that as the number of layers increases, the mutual information between each layer's embedding and the model input gradually decreases, while the mutual information between each layer's embedding and the model output gradually increases. For the decoding part, if the embeddings that is not closest to the output (behaviors) is used, then these embeddings might contain behaviorally irrelevant signals. Using these embeddings to generate behaviorally relevant signals could lead to the inclusion of irrelevant signals in the behaviorally relevant signals.
To demonstrate the above statement, we conducted experiments on the synthetic data. As shown in Table 2, we present the performance (neural R2 between the generated signals and the ground truth signals) of both models at several alpha values around the optimal alpha of dVAE (alpha=0.9) selected by the validation set. The experimental results show that at the same alpha value, the performance of d-VAE2 is consistently inferior to that of d-VAE, and d-VAE2 requires a higher alpha value to achieve performance comparable to d-VAE, and the best performance of d-VAE2 is inferior to that of d-VAE.
Author response table 3.
Neural R2 between generated signals and real behaviorally relevant signals
Thank you for your valuable feedback.
(1) Shwartz-Ziv, Ravid, and Naftali Tishby. "Opening the black box of deep neural networks via information." arXiv preprint arXiv:1703.00810 (2017).
Q7: “The beginning of the section on the "smaller R2 neurons" should clearly define what R2 is being discussed. Based on the response to previous reviewers, this R2 "signifies the proportion of neuronal activity variance explained by the linear encoding model, calculated using raw signals". This should be mentioned and made clear in the main text whenever this R2 is referred to.”
Thank you for your suggestion. We have made the modifications in the main text. Thank you for your valuable feedback.
Q8: “Various terms require clear definitions. The authors sometimes use vague terminology (e.g., "useless") without a clear definition. Similarly, discussions regarding dimensionality could benefit from more precise definitions. How is neural dimensionality defined? For example, how is "neural dimensionality of specific behaviors" (line 590) defined? Related to this, I agree with Reviewer 2 that a clear definition of irrelevant should be mentioned that clarifies that relevance is roughly taken as "correlated or predictive with a fixed time lag". The analyses do not explore relevance with arbitrary time lags between neural and behavior data.”
Thanks for your suggestion. We have removed the “useless” statements and have revised the statement of “the neural dimensionality of specific behaviors” in our revised manuscripts.
Regarding the use of fixed temporal lags, we followed the same practice as papers related to the dataset we use, which assume fixed temporal lags [1-3]. Furthermore, many studies in the motor cortex similarly use fixed temporal lags [4-6]. To clarify the definition, we have revised the definition in our manuscript. For details, please refer to the response to Q2 of reviewer #2 and our revised manuscript. We believe our definition is clearly articulated.
Thank you for your valuable feedback.
(1) Wang, Fang, et al. "Quantized attention-gated kernel reinforcement learning for brain– machine interface decoding." IEEE transactions on neural networks and learning systems 28.4 (2015): 873-886.
(2) Dyer, Eva L., et al. "A cryptography-based approach for movement decoding." Nature biomedical engineering 1.12 (2017): 967-976.
(3) Ahmadi, Nur, Timothy G. Constandinou, and Christos-Savvas Bouganis. "Robust and accurate decoding of hand kinematics from entire spiking activity using deep learning." Journal of Neural Engineering 18.2 (2021): 026011.
(4) Churchland, Mark M., et al. "Neural population dynamics during reaching." Nature 487.7405 (2012): 51-56.
(5) Kaufman, Matthew T., et al. "Cortical activity in the null space: permitting preparation without movement." Nature neuroscience 17.3 (2014): 440-448.
(6) Elsayed, Gamaleldin F., et al. "Reorganization between preparatory and movement population responses in motor cortex." Nature communications 7.1 (2016): 13239.
Q9: “CEBRA itself doesn't provide a neural reconstruction from its embeddings, but one could obtain one via a regression from extracted CEBRA embeddings to neural data. In addition to decoding results of CEBRA (figure S3), the neural reconstruction of CEBRA should be computed and CEBRA should be added to Figure 2 to see how the behaviorally relevant and irrelevant signals from CEBRA compare to other methods.”
Thank you for your question. Modifying CEBRA is beyond the scope of our work. As CEBRA is not a generative model, it cannot obtain behaviorally relevant and irrelevant signals, and therefore it lacks the results presented in Fig. 2. To avoid the same confusion encountered by reviewers #3 and #4 among our readers, we have opted to exclude the comparison with CEBRA. It is crucial to note, as previously stated, that our assessment of decoding capabilities has been benchmarked against the performance of the ANN on raw signals, which almost represents the upper limit of performance. Consequently, omitting CEBRA does not affect our conclusions.
Thank you for your valuable feedback.
Q10: “Line 923: "The optimal hyperparameter is selected based on the lowest averaged loss of five-fold training data." => why is this explained specifically under CEBRA? Isn't the same criteria used for hyperparameters of other methods? If so, clarify.”
Thank you for your question. The hyperparameter selection for CEBRA follows the practice of the original CEBRA paper. The hyperparameter selection for generative models is detailed in the Section “The strategy for selecting effective behaviorally-relevant signals”. Thank you for your valuable feedback.
-

-

Author Response
The following is the authors’ response to the previous reviews.
To the Senior Editor and the Reviewing Editor:
We sincerely appreciate the valuable comments provided by the reviewers, the reviewing editor, and the senior editor. After carefully reviewing and considering the comments, we have addressed the key concerns raised by the reviewers and made appropriate modifications to the article in the revised manuscript.
The main revisions made to the manuscript are as follows:
We have added comparison experiments with TNDM (see Fig. 2 and Fig. S2).
We conducted new synthetic experiments to demonstrate that our conclusions are not a by-product of d-VAE (see Fig. S2 and Fig. S11).
We have provided a detailed explanation of how our proposed criteria, especially the second criterion, can effectively exclude the selection of …
Author Response
The following is the authors’ response to the previous reviews.
To the Senior Editor and the Reviewing Editor:
We sincerely appreciate the valuable comments provided by the reviewers, the reviewing editor, and the senior editor. After carefully reviewing and considering the comments, we have addressed the key concerns raised by the reviewers and made appropriate modifications to the article in the revised manuscript.
The main revisions made to the manuscript are as follows:
We have added comparison experiments with TNDM (see Fig. 2 and Fig. S2).
We conducted new synthetic experiments to demonstrate that our conclusions are not a by-product of d-VAE (see Fig. S2 and Fig. S11).
We have provided a detailed explanation of how our proposed criteria, especially the second criterion, can effectively exclude the selection of unsuitable signals.
We have included a semantic overview figure of d-VAE (Fig. S1) and a visualization plot of latent variables (Fig. S13).
We have elaborated on the model details of d-VAE, as well as the hyperparameter selection and experimental settings of other comparison models.
We believe these revisions have significantly improved the clarity and comprehensibility of the manuscript. Thank you for the opportunity to address these important points.
Reviewer #1
Q1: “First, the model in the paper is almost identical to an existing VAE model (TNDM) that makes use of weak supervision with behaviour in the same way [1]. This paper should at least be referenced. If the authors wish they could compare their model to TNDM, which combines a state space model with smoothing similar to LFADS. Given that TNDM achieves very good behaviour reconstructions, it may be on par with this model without the need for a Kalman filter (and hence may achieve better separation of behaviour-related and unrelated dynamics).”
Our model significantly differs from TNDM in several aspects. While TNDM also constrains latent variables to decode behavioral information, it does not impose constraints to maximize behavioral information in the generated relevant signals. The trade-off between the decoding and reconstruction capabilities of generated relevant signals is the most significant contribution of our approach, which is not reflected in TNDM. In addition, the backbone network of signal extraction and the prior distribution of the two models are also different.
It's worth noting that our method does not require a Kalman filter. Kalman filter is used for post hoc assessment of the linear decoding ability of the generated signals. Please note that extracting and evaluating relevant signals are two distinct stages.
Heeding your suggestion, we have incorporated comparison experiments involving TNDM into the revised manuscript. Detailed information on model hyperparameters and training settings can be found in the Methods section in the revised manuscripts.
Thank you for your valuable feedback.
Q2: “Second, in my opinion, the claims regarding identifiability are overstated - this matters as the results depend on this to some extent. Recent work shows that VAEs generally suffer from identifiability problems due to the Gaussian latent space [2]. This paper also hints that weak supervision may help to resolve such issues, so this model as well as TNDM and CEBRA may indeed benefit from this. In addition however, it appears that the relative weight of the KL Divergence in the VAE objective is chosen very small compared to the likelihood (0.1%), so the influence of the prior is weak and the model may essentially learn the average neural trajectories while underestimating the noise in the latent variables. This, in turn, could mean that the model will not autoencode neural activity as well as it should, note that an average R2 in this case will still be high (I could not see how this is actually computed). At the same time, the behaviour R2 will be large simply because the different movement trajectories are very distinct. Since the paper makes claims about the roles of different neurons, it would be important to understand how well their single trial activities are reconstructed, which can perhaps best be investigated by comparing the Poisson likelihood (LFADS is a good baseline model). Taken together, while it certainly makes sense that well-tuned neurons contribute more to behaviour decoding, I worry that the very interesting claim that neurons with weak tuning contain behavioural signals is not well supported.”
We don’t think our distilled signals are average neural trajectories without variability. The quality of reconstructing single trial activities can be observed in Figure 3i and Figure S4. Neural trajectories in Fig. 3i and Fig. S4 show that our distilled signals are not average neural trajectories. Furthermore, if each trial activity closely matched the average neural trajectory, the Fano Factor (FF) should theoretically approach 0. However, our distilled signals exhibit a notable departure from this expectation, as evident in Figure 3c, d, g, and f. Regarding the diminished influence of the KL Divergence: Given that the ground truth of latent variable distribution is unknown, even a learned prior distribution might not accurately reflect the true distribution. We found the pronounced impact of the KL divergence would prove detrimental to the decoding and reconstruction performance. As a result, we opt to reduce the weight of the KL divergence term. Even so, KL divergence can still effectively align the distribution of latent variables with the distribution of prior latent variables, as illustrated in Fig. S13. Notably, our goal is extracting behaviorally-relevant signals from given raw signals rather than generating diverse samples from the prior distribution. When aim to separating relevant signals, we recommend reducing the influence of KL divergence. Regarding comparing the Poisson likelihood: We compared Poisson log-likelihood among different methods (except PSID since their obtained signals have negative values), and the results show that d-VAE outperforms other methods.
Author response image 1.
Regarding how R2 is computed:
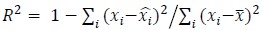 , where
, where 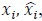 and
and 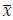 denote ith sample of raw signals, ith sample of distilled relevant signals, and the mean of raw signals. If the distilled signals exactly match the raw signals, the sum of squared error is zero, thus R2=1. If the distilled signals
denote ith sample of raw signals, ith sample of distilled relevant signals, and the mean of raw signals. If the distilled signals exactly match the raw signals, the sum of squared error is zero, thus R2=1. If the distilled signals 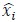 always are equal to
always are equal to 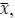 R2=0. If the distilled signals are worse than the mean estimation, R2 is negative, negative R2 is set to zero.
R2=0. If the distilled signals are worse than the mean estimation, R2 is negative, negative R2 is set to zero.Thank you for your valuable feedback.
Q3: “Third, and relating to this issue, I could not entirely follow the reasoning in the section arguing that behavioural information can be inferred from neurons with weak selectivity, but that it is not linearly decodable. It is right to test if weak supervision signals bleed into the irrelevant subspace, but I could not follow the explanations. Why, for instance, is the ANN decoder on raw data (I assume this is a decoder trained fully supervised) not equal in performance to the revenant distilled signals? Should a well-trained non-linear decoder not simply yield a performance ceiling? Next, if I understand correctly, distilled signals were obtained from the full model. How does a model perform trained only on the weakly tuned neurons? Is it possible that the subspaces obtained with the model are just not optimally aligned for decoding? This could be a result of limited identifiability or model specifics that bias reconstruction to averages (a well-known problem of VAEs). I, therefore, think this analysis should be complemented with tests that do not depend on the model.”
Regarding “Why, for instance, is the ANN decoder on raw data (I assume this is a decoder trained fully supervised) not equal in performance to the relevant distilled signals? Should a well-trained non-linear decoder not simply yield a performance ceiling?”: In fact, the decoding performance of raw signals with ANN is quite close to the ceiling. However, due to the presence of significant irrelevant signals in raw signals, decoding models like deep neural networks are more prone to overfitting when trained on noisy raw signals compared to behaviorally-relevant signals. Consequently, we anticipate that the distilled signals will demonstrate superior decoding generalization. This phenomenon is evident in Fig. 2 and Fig. S1, where the decoding performance of the distilled signals surpasses that of the raw signals, albeit not by a substantial margin.
Regarding “Next, if I understand correctly, distilled signals were obtained from the full model. How does a model perform trained only on the weakly tuned neurons? Is it possible that the subspaces obtained with the model are just not optimally aligned for decoding?”:Distilled signals (involving all neurons) are obtained by d-VAE. Subsequently, we use ANN to evaluate the performance of smaller and larger R2 neurons. Please note that separating and evaluating relevant signals are two distinct stages.
Regarding the reasoning in the section arguing that smaller R2 neurons encode rich information, we would like to provide a detailed explanation:
After extracting relevant signals through d-VAE, we specifically selected neurons characterized by smaller R2 values (Here, R2 signifies the proportion of neuronal activity variance explained by the linear encoding model, calculated using raw signals). Subsequently, we employed both KF and ANN to assess the decoding performance of these neurons. Remarkably, our findings revealed that smaller R2 neurons, previously believed to carry limited behavioral information, indeed encode rich information.
In a subsequent step, we employed d-VAE to exclusively distill the raw signals of these smaller R2 neurons (distinct from the earlier experiment where d-VAE processed signals from all neurons). We then employed KF and ANN to evaluate the distilled smaller R2 neurons. Interestingly, we observed that we could not attain the same richness of information solely through the use of these smaller R2 neurons.
Consequently, we put forth and tested two hypotheses: First, that larger R2 neurons introduce additional signals into the smaller R2 neurons that do not exist in the real smaller R2 neurons. Second, that larger R2 neurons aid in restoring the original appearance of impaired smaller R2 neurons. Our proposed criteria and synthetic experiments substantiate the latter scenario.
Thank you for your valuable feedback.
Q4: “Finally, a more technical issue to note is related to the choice to learn a non-parametric prior instead of using a conventional Gaussian prior. How is this implemented? Is just a single sample taken during a forward pass? I worry this may be insufficient as this would not sample the prior well, and some other strategy such as importance sampling may be required (unless the prior is not relevant as it weakly contributed to the ELBO, in which case this choice seems not very relevant). Generally, it would be useful to see visualisations of the latent variables to see how information about behaviour is represented by the model.”
Regarding "how to implement the prior?": Please refer to Equation 7 in the revised manuscript; we have added detailed descriptions in the revised manuscript.
Regarding "Generally, it would be useful to see visualizations of the latent variables to see how information about behavior is represented by the model.": Note that our focus is not on latent variables but on distilled relevant signals. Nonetheless, at your request, we have added the visualization of latent variables in the revised manuscript. Please see Fig. S13 for details.
Thank you for your valuable feedback.
Recommendations: “A minor point: the word 'distill' in the name of the model may be a little misleading - in machine learning the term refers to the construction of smaller models with the same capabilities.
It should be useful to add a schematic picture of the model to ease comparison with related approaches.”
In the context of our model's functions, it operates as a distillation process, eliminating irrelevant signals and retaining the relevant ones. Although the name of our model may be a little misleading, it faithfully reflects what our model does.
I have added a schematic picture of d-VAE in the revised manuscript. Please see Fig. S1 for details.
Thank you for your valuable feedback.
Reviewer #2
Q1: “Is the apparently increased complexity of encoding vs decoding so unexpected given the entropy, sparseness, and high dimensionality of neural signals (the "encoding") compared to the smoothness and low dimensionality of typical behavioural signals (the "decoding") recorded in neuroscience experiments? This is the title of the paper so it seems to be the main result on which the authors expect readers to focus. ”
We use the term "unexpected" due to the disparity between our findings and the prior understanding concerning neural encoding and decoding. For neural encoding, as we said in the Introduction, in previous studies, weakly-tuned neurons are considered useless, and smaller variance PCs are considered noise, but we found they encode rich behavioral information. For neural decoding, the nonlinear decoding performance of raw signals is significantly superior to linear decoding. However, after eliminating the interference of irrelevant signals, we found the linear decoding performance is comparable to nonlinear decoding. Rooted in these findings, which counter previous thought, we employ the term "unexpected" to characterize our observations.
Thank you for your valuable feedback.
Q2: “I take issue with the premise that signals in the brain are "irrelevant" simply because they do not correlate with a fixed temporal lag with a particular behavioural feature hand-chosen by the experimenter. As an example, the presence of a reward signal in motor cortex [1] after the movement is likely to be of little use from the perspective of predicting kinematics from time-bin to time-bin using a fixed model across trials (the apparent definition of "relevant" for behaviour here), but an entire sub-field of neuroscience is dedicated to understanding the impact of these reward-related signals on future behaviour. Is there method sophisticated enough to see the behavioural "relevance" of this brief, transient, post-movement signal? This may just be an issue of semantics, and perhaps I read too much into the choice of words here. Perhaps the authors truly treat "irrelevant" and "without a fixed temporal correlation" as synonymous phrases and the issue is easily resolved with a clarifying parenthetical the first time the word "irrelevant" is used. But I remain troubled by some claims in the paper which lead me to believe that they read more deeply into the "irrelevancy" of these components.”
In this paper, we employ terms like ‘behaviorally-relevant’ and ‘behaviorally-irrelevant’ only regarding behavioral variables of interest measured within a given task, such as arm kinematics during a motor control task. A similar definition can be found in the PSID[1].
Thank you for your valuable feedback.
[1] Sani, Omid G., et al. "Modeling behaviorally relevant neural dynamics enabled by preferential subspace identification." Nature Neuroscience 24.1 (2021): 140-149.
Q3: “The authors claim the "irrelevant" responses underpin an unprecedented neuronal redundancy and reveal that movement behaviors are distributed in a higher-dimensional neural space than previously thought." Perhaps I just missed the logic, but I fail to see the evidence for this. The neural space is a fixed dimensionality based on the number of neurons. A more sparse and nonlinear distribution across this set of neurons may mean that linear methods such as PCA are not effective ways to approximate the dimensionality. But ultimately the behaviourally relevant signals seem quite low-dimensional in this paper even if they show some nonlinearity may help.”
The evidence for the “useless” responses underpin an unprecedented neuronal redundancy is shown in Fig. 5a, d and Fig. S9a. Specifically, the sum of the decoding performance of smaller R2 neurons and larger R2 neurons is significantly greater than that of all neurons for relevant signals (red bar), demonstrating that movement parameters are encoded very redundantly in neuronal population. In contrast, we can not find this degree of neural redundancy in raw signals (purple bar).
The evidence for the “useless” responses reveal that movement behaviors are distributed in a higher-dimensional neural space than previously thought is shown in the left plot (involving KF decoding) of Fig. 6c, f and Fig. S9f. Specifically, the improvement of KF using secondary signals is significantly higher than using raw signals composed of the same number of dimensions as the secondary signals. These results demonstrate that these dimensions, spanning roughly from ten to thirty, encode much information, suggesting that behavioral information exists in a higher-dimensional subspace than anticipated from raw signals.
Thank you for your valuable feedback.
Q5: “there is an apparent logical fallacy that begins in the abstract and persists in the paper: "Surprisingly, when incorporating often-ignored neural dimensions, behavioral information can be decoded linearly as accurately as nonlinear decoding, suggesting linear readout is performed in motor cortex." Don't get me wrong: the equivalency of linear and nonlinear decoding approaches on this dataset is interesting, and useful for neuroscientists in a practical sense. However, the paper expends much effort trying to make fundamental scientific claims that do not feel very strongly supported. This reviewer fails to see what we can learn about a set of neurons in the brain which are presumed to "read out" from motor cortex. These neurons will not have access to the data analyzed here. That a linear model can be conceived by an experimenter does not imply that the brain must use a linear model. The claim may be true, and it may well be that a linear readout is implemented in the brain. Other work [2,3] has shown that linear readouts of nonlinear neural activity patterns can explain some behavioural features. The claim in this paper, however, is not given enough”
Due to the limitations of current observational methods and our incomplete understanding of brain mechanisms, it is indeed challenging to ascertain the specific data the brain acquires to generate behavior and whether it employs a linear readout. Conventionally, the neural data recorded in the motor cortex do encode movement behaviors and can be used to analyze neural encoding and decoding. Based on these data, we found that the linear decoder KF achieves comparable performance to that of the nonlinear decoder ANN on distilled relevant signals. This finding has undergone validation across three widely used datasets, providing substantial evidence. Furthermore, we conducted experiments on synthetic data to show that this conclusion is not a by-product of our model. In the revised manuscript, we added a more detailed description of this conclusion.
Thank you for your valuable feedback.
Q6: “Relatedly, I would like to note that the exercise of arbitrarily dividing a continuous distribution of a statistic (the "R2") based on an arbitrary threshold is a conceptually flawed exercise. The authors read too much into the fact that neurons which have a low R2 w.r.t. PDs have behavioural information w.r.t. other methods. To this reviewer, it speaks more about the irrelevance, so to speak, of the preferred direction metric than anything fundamental about the brain.”
We chose the R2 threshold in accordance with the guidelines provided in reference [1]. It's worth mentioning that this threshold does not exert any significant influence on the overall conclusions.
Thank you for your valuable feedback.
[1] Inoue, Y., Mao, H., Suway, S.B., Orellana, J. and Schwartz, A.B., 2018. Decoding arm speed during reaching. Nature communications, 9(1), p.5243.
Q7: “I am afraid I may be missing something, as I did not understand the fano factor analysis of Figure 3. In a sense the behaviourally relevant signals must have lower FF given they are in effect tied to the temporally smooth (and consistent on average across trials) behavioural covariates. The point of the original Churchland paper was to show that producing a behaviour squelches the variance; naturally these must appear in the behaviourally relevant components. A control distribution or reference of some type would possibly help here.”
We agree that including reference signals could provide more context. The Churchland paper said stimulus onset can lead to a reduction in neural variability. However, our experiment focuses specifically on the reaching process, and thus, we don't have comparative experiments involving different types of signals.
Thank you for your valuable feedback.
Q8: “The authors compare the method to LFADS. While this is a reasonable benchmark as a prominent method in the field, LFADS does not attempt to solve the same problem as d-VAE. A better and much more fair comparison would be TNDM [4], an extension of LFADS which is designed to identify behaviourally relevant dimensions.”
We have added the comparison experiments with TNDM in the revised manuscript (see Fig. 2 and Fig. S2). The details of model hyperparameters and training settings can be found in the Methods section in the revised manuscripts.
Thank you for your valuable feedback.
Reviewer #3
Q1.1: “TNDM: LFADS is not the best baseline for comparison. The authors should have compared with TNDM (Hurwitz et al. 2021), which is an extension of LFADS that (unlike LFADS) actually attempts to extract behaviorally relevant factors by adding a behavior term to the loss. The code for TNDM is also available on Github. LFADS is not even supervised by behavior and does not aim to address the problem that d-VAE aims to address, so it is not the most appropriate comparison. ”
We have added the comparison experiments with TNDM in the revised manuscript (see Fig. 2 and Fig. S2). The details of model hyperparameters and training settings can be found in the Methods section in the revised manuscripts.
Thank you for your valuable feedback.
Q1.2: “LFADS: LFADS is a sequential autoencoder that processes sections of data (e.g. trials). No explanation is given in Methods for how the data was passed to LFADS. Was the moving averaged smoothed data passed to LFADS or the raw spiking data (at what bin size)? Was a gaussian loss used or a poisson loss? What are the trial lengths used in each dataset, from which part of trials? For dataset C that has back-to-back reaches, was data chopped into segments? How long were these segments? Were the edges of segments overlapped and averaged as in (Keshtkaran et al. 2022) to avoid noisy segment edges or not? These are all critical details that are not explained. The same details would also be needed for a TNDM comparison (comment 1.1) since it has largely the same architecture as LFADS.
It is also critical to briefly discuss these fundamental differences between the inputs of methods in the main text. LFADS uses a segment of data whereas VAE methods just use one sample at a time. What does this imply in the results? I guess as long as VAEs outperform LFADS it is ok, but if LFADS outperforms VAEs in a given metric, could it be because it received more data as input (a whole segment)? Why was the factor dimension set to 50? I presume it was to match the latent dimension of the VAE methods, but is the LFADS factor dimension the correct match for that to make things comparable?
I am also surprised by the results. How do the authors justify LFADS having lower neural similarity (fig 2d) than VAE methods that operate on single time steps? LFADS is not supervised by behavior, so of course I don't expect it to necessarily outperform methods on behavior decoding. But all LFADS aims to do is to reconstruct the neural data so at least in this metric it should be able to outperform VAEs that just operate on single time steps? Is it because LFADS smooths the data too much? This is important to discuss and show examples of. These are all critical nuances that need to be discussed to validate the results and interpret them.”
Regarding “Was the moving averaged smoothed data passed to LFADS or the raw spiking data (at what bin size)? Was a gaussian loss used or a poisson loss?”: The data used by all models was applied to the same preprocessing procedure. That is, using moving averaged smoothed data with three bins, where the bin size is 100ms. For all models except PSID, we used a Poisson loss.
Regrading “What are the trial lengths used in each dataset, from which part of trials? For dataset C that has back-to-back reaches, was data chopped into segments? How long were these segments? Were the edges of segments overlapped and averaged as in (Keshtkaran et al. 2022) to avoid noisy segment edges or not?”:
For datasets A and B, a trial length of eighteen is set. Trials with lengths below the threshold are zero-padded, while trials exceeding the threshold are truncated to the threshold length from their starting point. In dataset A, there are several trials with lengths considerably longer than that of most trials. We found that padding all trials with zeros to reach the maximum length (32) led to poor performance. Consequently, we chose a trial length of eighteen, effectively encompassing the durations of most trials and leading to the removal of approximately 9% of samples. For dataset B (center-out), the trial lengths are relatively consistent with small variation, and the maximum length across all trials is eighteen. For dataset C, we set the trial length as ten because we observed the video of this paradigm and found that the time for completing a single trial was approximately one second. The segments are not overlapped.
Regarding “Why was the factor dimension set to 50? I presume it was to match the latent dimension of the VAE methods, but is the LFADS factor dimension the correct match for that to make things comparable?”: We performed a grid search for latent dimensions in {10,20,50} and found 50 is the best.
Regarding “I am also surprised by the results. How do the authors justify LFADS having lower neural similarity (fig 2d) than VAE methods that operate on single time steps? LFADS is not supervised by behavior, so of course I don't expect it to necessarily outperform methods on behavior decoding. But all LFADS aims to do is to reconstruct the neural data so at least in this metric it should be able to outperform VAEs that just operate on single time steps? Is it because LFADS smooths the data too much?”: As you pointed out, we found that LFADS tends to produce excessively smooth and consistent data, which can lead to a reduction in neural similarity.
Thank you for your valuable feedback.
Q1.3: “PSID: PSID is linear and uses past input samples to predict the next sample in the output. Again, some setup choices are not well justified, and some details are left out in the 1-line explanation given in Methods.
Why was a latent dimension of 6 chosen? Is this the behaviorally relevant latent dimension or the total latent dimension (for the use case here it would make sense to set all latent states to be behaviorally relevant)? Why was a horizon hyperparameter of 3 chosen? First, it is important to mention fundamental parameters such as latent dimension for each method in the main text (not just in methods) to make the results interpretable. Second, these hyperparameters should be chosen with a grid search in each dataset (within the training data, based on performance on the validation part of the training data), just as the authors do for their method (line 779). Given that PSID isn't a deep learning method, doing a thorough grid search in each fold should be quite feasible. It is important that high values for latent dimension and a wider range of other hyperparmeters are included in the search, because based on how well the residuals (x_i) for this method are shown predict behavior in Fig 2, the method seems to not have been used appropriately. I would expect ANN to improve decoding for PSID versus its KF decoding since PSID is fully linear, but I don't expect KF to be able to decode so well using the residuals of PSID if the method is used correctly to extract all behaviorally relevant information from neural data. The low neural reconstruction in Fid 2d could also partly be due to using too small of a latent dimension.
Again, another import nuance is the input to this method and how differs with the input to VAE methods. The learned PSID model is a filter that operates on all past samples of input to predict the output in the "next" time step. To enable a fair comparison with VAE methods, the authors should make sure that the last sample "seen" by PSID is the same as then input sample seen by VAE methods. This is absolutely critical given how large the time steps are, otherwise PSID might underperform simply because it stopped receiving input 300ms earlier than the input received by VAE methods. To fix this, I think the authors can just shift the training and testing neural time series of PSID by 1 sample into the past (relative to the behavior), so that PSID's input would include the input of VAE methods. Otherwise, VAEs outperforming PSID is confounded by PSID's input not including the time step that was provided to VAE.”
Thanks for your suggestions for letting PSID see the current neural observations. We did it per your suggestions and then performed a grid search for the hyperparameters for PSID. Specifically, we performed a grid search for the horizon hyperparameter in {2,3,4,5,6,7}. Since the relevant latent dimension should be lower than the horizon times the dimension of behavior variables (two-dimensional velocity in this paper) and increasing the dimension will reach performance saturation, we directly set the relevant latent dimensions as the maximum. The horizon number of datasets A, B, C, and synthetic datasets is 7, 6, 6 and 5, respectively.
And thus the latent dimension of datasets A, B, and C and the synthetic dataset is 14, 12, 12 and 10, respectively.
Our experiments show that KF can decode information from irrelevant signals obtained by PSID. Although PSID extracts the linear part of raw signals, KF can still use the linear part of the residuals for decoding. The low reconstruction performance of PSID may be because the relationship between latent variables and neural signals is linear, and the relationship between latent variables and behaviors is also linear; this is equivalent to the linear relationship between behaviors and neural signals, and linear models can only explain a small fraction of neural signals.
Thank you for your valuable feedback.
Q1.4: “CEBRA: results for CEBRA are incomplete. Similarity to raw signals is not shown. Decoding of behaviorally irrelevant residuals for CEBRA is not shown. Per Fig. S2, CEBRA does better or similar ANN decoding in datasets A and C, is only slightly worse in Dataset B, so it is important to show the other key metrics otherwise it is unclear whether d-VAE has some tangible advantage over CEBRA in those 2 datasets or if they are similar in every metric. Finally, it would be better if the authors show the results for CEBRA on Fig. 2, just as is done for other methods because otherwise it is hard to compare all methods.”
CEBRA is a non-generative model, this model cannot generate behaviorally-relevant signals. Therefore, we only compared the decoding performance of latent embeddings of CEBRA and signals of d-VAE.
Thank you for your valuable feedback.
Q2: “Given the fact that d-VAE infers the latent (z) based on the population activity (x), claims about properties of the inferred behaviorally relevant signals (x_r) that attribute properties to individual neurons are confounded.
The authors contrast their approach to population level approaches in that it infers behaviorally relevant signals for individual neurons. However, d-VAE is also a population method as it aggregates population information to infer the latent (z), from which behaviorally relevant part of the activity of each neuron (x_r) is inferred. The authors note this population level aggregation of information as a benefit of d-VAE, but only acknowledge it as a confound briefly in the context of one of their analyses (line 340): "The first is that the larger R2 neurons leak their information to the smaller R2 neurons, causing them contain too much behavioral information". They go on to dismiss this confounding possibility by showing that the inferred behaviorally relevant signal of each neuron is often most similar to its own raw signals (line 348-352) compared with all other neurons. They also provide another argument specific to that result section (i.e., residuals are not very behavior predictive), which is not general so I won't discuss it in depth here. These arguments however do not change the basic fact that d-VAE aggregates information from other neurons when extracting the behaviorally relevant activity of any given neuron, something that the authors note as a benefit of d-VAE in many instances. The fact that d-VAE aggregates population level info to give the inferred behaviorally relevant signal for each neuron confounds several key conclusions. For example, because information is aggregated across neurons, when trial to trial variability looks smoother after applying d-VAE (Fig 3i), or reveals better cosine tuning (Fig 3b), or when neurons that were not very predictive of behavior become more predictive of behavior (Fig 5), one cannot really attribute the new smoother single trial activity or the improved decoding to the same single neurons; rather these new signals/performances include information from other neurons. Unless the connections of the encoder network (z=f(x)) is zero for all other neurons, one cannot claim that the inferred rates for the neuron are truly solely associated with that neuron. I believe this a fundamental property of a population level VAE, and simply makes the architecture unsuitable for claims regarding inherent properties of single neurons. This confound is partly why the first claim in the abstract are not supported by data: observing that neurons that don't predict behavior very well would predict it much better after applying d-VAE does not prove that these neurons themselves "encode rich[er] behavioral information in complex nonlinear ways" (i.e., the first conclusion highlighted in the abstract) because information was also aggregated from other neurons. The other reason why this claim is not supported by data is the characterization of the encoding for smaller R2 neurons as "complex nonlinear", which the method is not well equipped to tease apart from linear mappings as I explain in my comment 3.”
We acknowledge that we cannot obtain the exact single neuronal activity that does not contain any information from other neurons. However, we believe our model can extract accurate approximation signals of the ground truth relevant signals. These signals preserve the inherent properties of single neuronal activity to some extent and can be used for analysis at the single-neuron level.
We believe d-VAE is a reasonable approach to extract effective relevant signals that preserve inherent properties of single neuronal activity for four key reasons:
d-VAE is a latent variable model that adheres to the neural population doctrine. The neural population doctrine posits that information is encoded within interconnected groups of neurons, with the existence of latent variables (neural modes) responsible for generating observable neuronal activity [1, 2]. If we can perfectly obtain the true generative model from latent variables to neuronal activity, then we can generate the activity of each neuron from hidden variables without containing any information from other neurons. However, without a complete understanding of the brain’s encoding strategies (or generative model), we can only get the approximation signals of the ground truth signals.
After the generative model is established, we need to infer the parameters of the generative model and the distribution of latent variables. During the inference process, inference algorithms such as variational inference or EM algorithms will be used. Generally, the obtained latent variables are also approximations of the real latent variables. When inferring the latent variables, it is inevitable to aggregation the information of the neural population, and latent variables are derived through weighted combinations of neuronal populations [3].
This inference process is consistent with that of d-VAE (or VAE-based models).
- Latent variables are derived from raw neural signals and used to explain raw neural signals. Considering the unknown ground truth of latent variables and behaviorally-relevant signals, it becomes evident that the only reliable reference at the signal level is the raw signals. A crucial criterion for evaluating the reliability of latent variable models (including latent variables and generated relevant signals) is their capability to effectively explain the raw signals [3]. Consequently, we firmly maintain the belief that if the generated signals closely resemble the raw signals to the greatest extent possible, in accordance with an equivalence principle, we can claim that these obtained signals faithfully retain the inherent properties of single neurons. d-VAE explicitly constrains the generated signal to closely resemble the raw signals. These results demonstrate that d-VAE can extract effective relevant signals that preserve inherent properties of single neuronal activity.
Based on the above reasons, we hold that generating single neuronal activities with the VAE framework is a reasonable approach. The remaining question is whether our model can obtain accurate relevant signals in the absence of ground truth. To our knowledge, in cases where the ground truth of relevant signals is unknown, there are typically two approaches to verifying the reliability of extracted signals:
Conducting synthetic experiments where the ground truth is known.
Validation based on expert knowledge (Three criteria were proposed in this paper). Both our extracted signals and key conclusions have been validated using these two approaches.
Next, we will provide a detailed response to the concerns regarding our first key conclusion that smaller R2 neurons encode rich information.
We acknowledge that larger R2 neurons play a role in aiding the reconstruction of signals in smaller R2 neurons through their neural activity. However, considering that neurons are correlated rather than independent entities, we maintain the belief that larger R2 neurons assist damaged smaller R2 neurons in restoring their original appearance. Taking image denoising as an example, when restoring noisy pixels to their original appearance, relying solely on the noisy pixels themselves is often impractical. Assistance from their correlated, clean neighboring pixels becomes necessary.
The case we need to be cautious of is that the larger R2 neurons introduce additional signals (m) that contain substantial information to smaller R2 neurons, which they do not inherently possess. We believe this case does not hold for two reasons. Firstly, logically, adding extra signals decreases the reconstruction performance, and the information carried by these additional signals is redundant for larger R2 neurons, thus they do not introduce new information that can enhance the decoding performance of the neural population. Therefore, it seems unlikely and unnecessary for neural networks to engage in such counterproductive actions. Secondly, even if this occurs, our second criterion can effectively exclude the selection of these signals. To clarify, if we assume that x, y, and z denote the raw, relevant, and irrelevant signals of smaller R2 neurons, with x=y+z, and the extracted relevant signals become y+m, the irrelevant signals become z-m in this case. Consequently, the irrelevant signals contain a significant amount of information. It's essential to emphasize that this criterion holds significant importance in excluding undesirable signals.
Furthermore, we conducted a synthetic experiment to show that d-VAE can indeed restore the damaged information of smaller R2 neurons with the help of larger R2 neurons, and the restored neuronal activities are more similar to ground truth compared to damaged raw signals. Please see Fig. S11a,b for details.
Thank you for your valuable feedback.
[1] Saxena, S. and Cunningham, J.P., 2019. Towards the neural population doctrine. Current opinion in neurobiology, 55, pp.103-111.
[2] Gallego, J.A., Perich, M.G., Miller, L.E. and Solla, S.A., 2017. Neural manifolds for the control of movement. Neuron, 94(5), pp.978-984.
[3] Cunningham, J.P. and Yu, B.M., 2014. Dimensionality reduction for large-scale neural recordings. Nature neuroscience, 17(11), pp.1500-1509.
Q3: “Given the nonlinear architecture of the VAE, claims about the linearity or nonlinearity of cortical readout are confounded and not supported by the results.
The inference of behaviorally relevant signals from raw signals is a nonlinear operation, that is x_r=g(f(x)) is nonlinear function of x. So even when a linear KF is used to decode behavior from the inferred behaviorally relevant signals, the overall decoding from raw signals to predicted behavior (i.e., KF applied to g(f(x))) is nonlinear. Thus, the result that decoding of behavior from inferred behaviorally relevant signals (x_r) using a linear KF and a nonlinear ANN reaches similar accuracy (Fig 2), does not suggest that a "linear readout is performed in the motor cortex", as the authors claim (line 471). The authors acknowledge this confound (line 472) but fail to address it adequately. They perform a simulation analysis where the decoding gap between KF and ANN remains unchanged even when d-VAE is used to infer behaviorally relevant signals in the simulation. However, this analysis is not enough for "eliminating the doubt" regarding the confound. I'm sure the authors can also design simulations where the opposite happens and just like in the data, d-VAE can improve linear decoding to match ANN decoding. An adequate way to address this concern would be to use a fully linear version of the autoencoder where the f(.) and g(.) mappings are fully linear. They can simply replace these two networks in their model with affine mappings, redo the modeling and see if the model still helps the KF decoding accuracy reach that of the ANN decoding. In such a scenario, because the overall KF decoding from original raw signals to predicted behavior (linear d-VAE + KF) is linear, then they could move toward the claim that the readout is linear. Even though such a conclusion would still be impaired by the nonlinear reference (d-VAE + ANN decoding) because the achieved nonlinear decoding performance could always be limited by network design and fitting issues. Overall, the third conclusion highlighted in the abstract is a very difficult claim to prove and is unfortunately not supported by the results.”
We aim to explore the readout mechanism of behaviorally-relevant signals, rather than raw signals. Theoretically, the process of removing irrelevant signals should not be considered part of the inherent decoding mechanisms of the relevant signals. Assuming that the relevant signals we extracted are accurate, the conclusion of linear readout is established. On the synthetic data where the ground truth is known, our distilled signals show a significant improvement in neural similarity to the ground truth when compared to raw signals (refer to Fig. S2l). This observation demonstrates that our distilled signals are accurate approximations of the ground truth. Furthermore, on the three widely-used real datasets, our distilled signals meet the stringent criteria we have proposed (see Fig. 2), also providing strong evidence for their accuracy.
Regarding the assertion that we could create simulations in which d-VAE can make signals that are inherently nonlinearly decodable into linearly decodable ones: In reality, we cannot achieve this, as the second criterion can rule out the selection of such signals. Specifically,z=x+y=n^2+y, where z, x, y, and n denote raw signals, relevant signals, irrelevant signals and latent variables. If the relevant signals obtained by d-VAE are n, then these signals can be linear decoded accurately. However, the corresponding irrelevant signals are n^2-n+z; thus, irrelevant signals will have much information, and these extracted relevant signals will not be selected. Furthermore, our synthetic experiments offer additional evidence supporting the conclusion that d-VAE does not make inherently nonlinearly decodable signals become linearly decodable ones. As depicted in Fig. S11c, there exists a significant performance gap between KF and ANN when decoding the ground truth signals of smaller R2 neurons. KF exhibits notably low performance, leaving substantial room for compensation by d-VAE. However, following processing by d-VAE, KF's performance of distilled signals fails to surpass its already low ground truth performance and remains significantly inferior to ANN's performance. These results collectively confirm that our approach does not convert signals that are inherently nonlinearly decodable into linearly decodable ones, and the conclusion of linear readout is not a by-product by d-VAE.
Regarding the suggestion of using linear d-VAE + KF, as discussed in the Discussion section, removing the irrelevant signals requires a nonlinear operation, and linear d-VAE can not effectively separate relevant and irrelevant signals.
Thank you for your valuable feedback.
Q4: “The authors interpret several results as indications that "behavioral information is distributed in a higher-dimensional subspace than expected from raw signals", which is the second main conclusion highlighted in the abstract. However, several of these arguments do not convincingly support that conclusion.
4.1) The authors observe that behaviorally relevant signals for neurons with small principal components (referred to as secondary) have worse decoding with KF but better decoding with ANN (Fig. 6b,e), which also outperforms ANN decoding from raw signals. This observation is taken to suggest that these secondary behaviorally relevant signals encode behavior information in highly nonlinear ways and in a higher dimensions neural space than expected (lines 424 and 428). These conclusions however are confounded by the fact that A) d-VAE uses nonlinear encoding, so one cannot conclude from ANN outperforming KF that behavior is encoded nonlinearly in the motor cortex (see comment 3 above), and B) d-VAE aggregates information across the population so one cannot conclude that these secondary neurons themselves had as much behavior information (see comment 2 above).
4.2) The authors observe that the addition of the inferred behaviorally relevant signals for neurons with small principal components (referred to as secondary) improves the decoding of KF more than it improves the decoding of ANN (red curves in Fig 6c,f). This again is interpreted similarly as in 4.1, and is confounded for similar reasons (line 439): "These results demonstrate that irrelevant signals conceal the smaller variance PC signals, making their encoded information difficult to be linearly decoded, suggesting that behavioral information exists in a higher-dimensional subspace than anticipated from raw signals". This is confounded by because of the two reasons explained in 4.1. To conclude nonlinear encoding based on the difference in KF and ANN decoding, the authors would need to make the encoding/decoding in their VAE linear to have a fully linear decoder on one hand (with linear d-VAE + KF) and a nonlinear decoder on the other hand (with linear d-VAE + ANN), as explained in comment 3.
4.3) From S Fig 8, where the authors compare cumulative variance of PCs for raw and inferred behaviorally relevant signals, the authors conclude that (line 554): "behaviorally-irrelevant signals can cause an overestimation of the neural dimensionality of behaviorally-relevant responses (Supplementary Fig. S8)." However, this analysis does not really say anything about overestimation of "behaviorally relevant" neural dimensionality since the comparison is done with the dimensionality of "raw" signals. The next sentence is ok though: "These findings highlight the need to filter out relevant signals when estimating the neural dimensionality.", because they use the phrase "neural dimensionality" not "neural dimensionality of behaviorally-relevant responses".”
Questions 4.1 and 4.2 are a combination of Q2 and Q3. Please refer to our responses to Q2 and Q3.
Regarding question 4.3 about “behaviorally-irrelevant signals can cause an overestimation of the neural dimensionality of behaviorally-relevant responses”: Previous studies usually used raw signals to estimate the neural dimensionality of specific behaviors. We mean that using raw signals, which include many irrelevant signals, will cause an overestimation of the neural dimensionality. We have modified this sentence in the revised manuscripts.
Thank you for your valuable feedback.
Q5: “Imprecise use of language in many places leads to inaccurate statements. I will list some of these statements”
5.1) In the abstract: "One solution is to accurately separate behaviorally-relevant and irrelevant signals, but this approach remains elusive due to the unknown ground truth of behaviorally-relevant signals". This statement is not accurate because it implies no prior work does this. The authors should make their statement more specific and also refer to some goal that existing linear (e.g., PSID) and nonlinear (e.g., TNDM) methods for extracting behaviorally relevant signals fail to achieve.
5.2) In the abstract: "we found neural responses previously considered useless encode rich behavioral information" => what does "useless" mean operationally? Low behavior tuning? More precise use of language would be better.
5.3) "... recent studies (Glaser 58 et al., 2020; Willsey et al., 2022) demonstrate nonlinear readout outperforms linear readout." => do these studies show that nonlinear "readout" outperforms linear "readout", or just that nonlinear models outperform linear models?
5.4) Line 144: "The first criterion is that the decoding performance of the behaviorally-relevant signals (red bar, Fig.1) should surpass that of raw signals (the red dotted line, Fig.1).". Do the authors mean linear decoding here or decoding in general? If the latter, how can something extracted from neural surpass decoding of neural data, when the extraction itself can be thought of as part of decoding? The operational definition for this "decoding performance" should be clarified.
5.5) Line 311: "we found that the dimensionality of primary subspace of raw signals (26, 64, and 45 for datasets A, B, and C) is significantly higher than that of behaviorally-relevant signals (7, 13, and 9), indicating that behaviorally-irrelevant signals lead to an overestimation of the neural dimensionality of behaviorally-relevant signals." => here the dimensionality of the total PC space (i.e., primary subspace of raw signals) is being compared with that of inferred behaviorally-relevant signals, so the former being higher does not indicate that neural dimensionality of behaviorally-relevant signals was overestimated. The former is simply not behavioral so this conclusion is not accurate.
5.6) Section "Distilled behaviorally-relevant signals uncover that smaller R2 neurons encode rich behavioral information in complex nonlinear ways". Based on what kind of R2 are the neurons grouped? Behavior decoding R2 from raw signals? Using what mapping? Using KF? If KF is used, the result that small R2 neurons benefit a lot from d-VAE could be somewhat expected, given the nonlinearity of d-VAE: because only ANN would have the capacity to unwrap the nonlinear encoding of d-VAE as needed. If decoding performance that is used to group neurons is based on data, regression to the mean could also partially explain the result: the neurons with worst raw decoding are most likely to benefit from a change in decoder, than neurons that already had good decoding. In any case, the R2 used to partition and sort neurons should be more clearly stated and reminded throughout the text and I Fig 3.
5.7) Line 346 "...it is impossible for our model to add the activity of larger R2 neurons to that of smaller R2 neurons" => Is it really impossible? The optimization can definitely add small-scale copies of behaviorally relevant information to all neurons with minimal increase in the overall optimization loss, so this statement seems inaccurate.
5.8) Line 490: "we found that linear decoders can achieve comparable performance to that of nonlinear decoders, providing compelling evidence for the presence of linear readout in the motor cortex." => inaccurate because no d-VAE decoding is really linear, as explained in comment 3 above.
5.9) Line 578: ". However, our results challenge this idea by showing that signals composed of smaller variance PCs nonlinearly encode a significant amount of behavioral information." => inaccurate as results are confounded by nonlinearity of d-VAE as explained in comment 3 above.
5.10) Line 592: "By filtering out behaviorally-irrelevant signals, our study found that accurate decoding performance can be achieved through linear readout, suggesting that the motor cortex may perform linear readout to generate movement behaviors." => inaccurate because it us confounded by the nonlinearity of d-VAE as explained in comment 3 above.”
Regarding “5.1) In the abstract: "One solution is to accurately separate behaviorally-relevant and irrelevant signals, but this approach remains elusive due to the unknown ground truth of behaviorally-relevant signals". This statement is not accurate because it implies no prior work does this. The authors should make their statement more specific and also refer to some goal that existing linear (e.g., PSID) and nonlinear (e.g., TNDM) methods for extracting behaviorally relevant signals fail to achieve”:
We believe our statement is accurate. Our primary objective is to extract accurate behaviorally-relevant signals that closely approximate the ground truth relevant signals. To achieve this, we strike a balance between the reconstruction and decoding performance of the generated signals, aiming to effectively capture the relevant signals. This crucial aspect of our approach sets it apart from other methods. In contrast, other methods tend to emphasize the extraction of valuable latent neural dynamics. We have provided elaboration on the distinctions between d-VAE and other approaches in the Introduction and Discussion sections.
Thank you for your valuable feedback.
Regarding “5.2) In the abstract: "we found neural responses previously considered useless encode rich behavioral information" => what does "useless" mean operationally? Low behavior tuning? More precise use of language would be better.”:
In the analysis of neural signals, smaller variance PC signals are typically seen as noise and are often discarded. Similarly, smaller R2 neurons are commonly thought to be dominated by noise and are not further analyzed. Given these considerations, we believe that the term "considered useless" is appropriate in this context. Thank you for your valuable feedback.
Regarding “5.3) "... recent studies (Glaser 58 et al., 2020; Willsey et al., 2022) demonstrate nonlinear readout outperforms linear readout." => do these studies show that nonlinear "readout" outperforms linear "readout", or just that nonlinear models outperform linear models?”:
In this paper, we consider the two statements to be equivalent. Thank you for your valuable feedback.
Regarding “5.4) Line 144: "The first criterion is that the decoding performance of the behaviorally-relevant signals (red bar, Fig.1) should surpass that of raw signals (the red dotted line, Fig.1).". Do the authors mean linear decoding here or decoding in general? If the latter, how can something extracted from neural surpass decoding of neural data, when the extraction itself can be thought of as part of decoding? The operational definition for this "decoding performance" should be clarified.”:
We mean the latter, as we said in the section “Framework for defining, extracting, and separating behaviorally-relevant signals”, since raw signals contain too many behaviorally-irrelevant signals, deep neural networks are more prone to overfit raw signals than relevant signals. Therefore the decoding performance of relevant signals should surpass that of raw signals. Thank you for your valuable feedback.
Regarding “5.5) Line 311: "we found that the dimensionality of primary subspace of raw signals (26, 64, and 45 for datasets A, B, and C) is significantly higher than that of behaviorally-relevant signals (7, 13, and 9), indicating that behaviorally-irrelevant signals lead to an overestimation of the neural dimensionality of behaviorally-relevant signals." => here the dimensionality of the total PC space (i.e., primary subspace of raw signals) is being compared with that of inferred behaviorally-relevant signals, so the former being higher does not indicate that neural dimensionality of behaviorally-relevant signals was overestimated. The former is simply not behavioral so this conclusion is not accurate.”: In practice, researchers usually used raw signals to estimate the neural dimensionality. We mean that using raw signals to do this would overestimate the neural dimensionality. Thank you for your valuable feedback.
Regarding “5.6) Section "Distilled behaviorally-relevant signals uncover that smaller R2 neurons encode rich behavioral information in complex nonlinear ways". Based on what kind of R2 are the neurons grouped? Behavior decoding R2 from raw signals? Using what mapping? Using KF? If KF is used, the result that small R2 neurons benefit a lot from d-VAE could be somewhat expected, given the nonlinearity of d-VAE: because only ANN would have the capacity to unwrap the nonlinear encoding of d-VAE as needed. If decoding performance that is used to group neurons is based on data, regression to the mean could also partially explain the result: the neurons with worst raw decoding are most likely to benefit from a change in decoder, than neurons that already had good decoding. In any case, the R2 used to partition and sort neurons should be more clearly stated and reminded throughout the text and I Fig 3.”:
When employing R2 to characterize neurons, it indicates the extent to which neuronal activity is explained by the linear encoding model [1-3]. Smaller R2 neurons have a lower capacity for linearly tuning (encoding) behaviors, while larger R2 neurons have a higher capacity for linearly tuning (encoding) behaviors. Specifically, the approach involves first establishing an encoding relationship from velocity to neural signal using a linear model, i.e., y=f(x), where f represents a linear regression model, x denotes velocity, and y denotes the neural signal. Subsequently, R2 is utilized to quantify the effectiveness of the linear encoding model in explaining neural activity. We have provided a comprehensive explanation in the revised manuscript. Thank you for your valuable feedback.
[1] Collinger, J.L., Wodlinger, B., Downey, J.E., Wang, W., Tyler-Kabara, E.C., Weber, D.J., McMorland, A.J., Velliste, M., Boninger, M.L. and Schwartz, A.B., 2013. High-performance neuroprosthetic control by an individual with tetraplegia. The Lancet, 381(9866), pp.557-564.
[2] Wodlinger, B., et al. "Ten-dimensional anthropomorphic arm control in a human brain− machine interface: difficulties, solutions, and limitations." Journal of neural engineering 12.1 (2014): 016011.
[3] Inoue, Y., Mao, H., Suway, S.B., Orellana, J. and Schwartz, A.B., 2018. Decoding arm speed during reaching. Nature communications, 9(1), p.5243.
Regarding Questions 5.7, 5.8, 5.9, and 5.10:
We believe our conclusions are solid. The reasons can be found in our replies in Q2 and Q3. Thank you for your valuable feedback.
Q6: “Imprecise use of language also sometimes is not inaccurate but just makes the text hard to follow.
6.1) Line 41: "about neural encoding and decoding mechanisms" => what is the definition of encoding/decoding and how do these differ? The definitions given much later in line 77-79 is also not clear.
6.2) Line 323: remind the reader about what R2 is being discussed, e.g., R2 of decoding behavior using KF. It is critical to know if linear or nonlinear decoding is being discussed.
6.3) Line 488: "we found that neural responses previously considered trivial encode rich behavioral information in complex nonlinear ways" => "trivial" in what sense? These phrases would benefit from more precision, for example: "neurons that may seem to have little or no behavior information encoded". The same imprecise word ("trivial") is also used in many other places, for example in the caption of Fig S9.
6.4) Line 611: "The same should be true for the brain." => Too strong of a statement for an unsupported claim suggesting the brain does something along the lines of nonlin VAE + linear readout.
6.5) In Fig 1, legend: what is the operational definition of "generating performance"? Generating what? Neural reconstruction?”
Regarding “6.1) Line 41: "about neural encoding and decoding mechanisms" => what is the definition of encoding/decoding and how do these differ? The definitions given much later in line 77-79 is also not clear.”:
We would like to provide a detailed explanation of neural encoding and decoding. Neural encoding means how neuronal activity encodes the behaviors, that is, y=f(x), where y denotes neural activity and, x denotes behaviors, f is the encoding model. Neural decoding means how the brain decodes behaviors from neural activity, that is, x=g(y), where g is the decoding model. For further elaboration, please refer to [1]. We have included references that discuss the concepts of encoding and decoding in the revised manuscript. Thank you for your valuable feedback.
[1] Kriegeskorte, Nikolaus, and Pamela K. Douglas. "Interpreting encoding and decoding models." Current opinion in neurobiology 55 (2019): 167-179.
Regarding “6.2) Line 323: remind the reader about what R2 is being discussed, e.g., R2 of decoding behavior using KF. It is critical to know if linear or nonlinear decoding is being discussed.”:
This question is the same as Q5.6. Please refer to the response to Q5.6. Thank you for your valuable feedback.
Regarding “6.3) Line 488: "we found that neural responses previously considered trivial encode rich behavioral information in complex nonlinear ways" => "trivial" in what sense? These phrases would benefit from more precision, for example: "neurons that may seem to have little or no behavior information encoded". The same imprecise word ("trivial") is also used in many other places, for example in the caption of Fig S9.”:
We have revised this statement in the revised manuscript. Thanks for your recommendation.
Regarding “6.4) Line 611: "The same should be true for the brain." => Too strong of a statement for an unsupported claim suggesting the brain does something along the lines of nonlin VAE + linear readout.”
We mean that removing the interference of irrelevant signals and decoding the relevant signals should logically be two stages. We have revised this statement in the revised manuscript. Thank you for your valuable feedback.
Regarding “6.5) In Fig 1, legend: what is the operational definition of "generating performance"? Generating what? Neural reconstruction?””:
We have replaced “generating performance” with “reconstruction performance” in the revised manuscript. Thanks for your recommendation.
Q7: “In the analysis presented starting in line 449, the authors compare improvement gained for decoding various speed ranges by adding secondary (small PC) neurons to the KF decoder (Fig S11). Why is this done using the KF decoder, when earlier results suggest an ANN decoder is needed for accurate decoding from these small PC neurons? It makes sense to use the more accurate nonlinear ANN decoder to support the fundamental claim made here, that smaller variance PCs are involved in regulating precise control”
Because when the secondary signal is superimposed on the primary signal, the enhancement in KF performance is substantial. We wanted to explore in which aspect of the behavior the KF performance improvement is mainly reflected. In comparison, the improvement of ANN by the secondary signal is very small, rendering the exploration of the aforementioned questions inconsequential. Thank you for your valuable feedback.
Q8: “A key limitation of the VAE architecture is that it doesn't aggregate information over multiple time samples. This may be why the authors decided to use a very large bin size of 100ms and beyond that smooth the data with a moving average. This limitation should be clearly stated somewhere in contrast with methods that can aggregate information over time (e.g., TNDM, LFADS, PSID) ”
We have added this limitation in the Discussion in the revised manuscript. Thanks for your recommendation.
Q9: “Fig 5c and parts of the text explore the decoding when some neurons are dropped. These results should come with a reminder that dropping neurons from behaviorally relevant signals is not technically possible since the extraction of behaviorally relevant signals with d-VAE is a population level aggregation that requires the raw signal from all neurons as an input. This is also important to remind in some places in the text for example:
- Line 498: "...when one of the neurons is destroyed."
- Line 572: "In contrast, our results show that decoders maintain high performance on distilled signals even when many neurons drop out."”
We want to explore the robustness of real relevant signals in the face of neuron drop-out. The signals our model extracted are an approximation of the ground truth relevant signals and thus serve as a substitute for ground truth to study this problem. Thank you for your valuable feedback.
Q10: “Besides the confounded conclusions regarding the readout being linear (see comment 3 and items related to it in comment 5), the authors also don't adequately discuss prior works that suggest nonlinearity helps decoding of behavior from the motor cortex. Around line 594, a few works are discussed as support for the idea of a linear readout. This should be accompanied by a discussion of works that support a nonlinear encoding of behavior in the motor cortex, for example (Naufel et al. 2019; Glaser et al. 2020), some of which the authors cite elsewhere but don't discuss here.”
We have added this discussion in the revised manuscript. Thanks for your recommendation.
Q11: “Selection of hyperparameters is not clearly explained. Starting line 791, the authors give some explanation for one hyperparameter, but not others. How are the other hyperparameters determined? What is the search space for the grid search of each hyperparameter? Importantly, if hyperparameters are determined only based on the training data of each fold, why is only one value given for the hyperparameter selected in each dataset (line 814)? Did all 5 folds for each dataset happen to select exactly the same hyperparameter based on their 5 different training/validation data splits? That seems unlikely.”
We perform a grid search in {0.001, 0.01,0.1,1} for hyperparameter beta. And we found that 0.001 is the best for all datasets. As for the model parameters, such as hidden neuron numbers, this model capacity has reached saturation decoding performance and does not influence the results.
Regarding “Importantly, if hyperparameters are determined only based on the training data of each fold, why is only one value given for the hyperparameter selected in each dataset (line 814)? Did all 5 folds for each dataset happen to select exactly the same hyperparameter based on their 5 different training/validation data splits”: We selected the hyperparameter based on the average performance of 5 folds data on validation sets. The selected value denotes the one that yields the highest average performance across the 5 folds data.
Thank you for your valuable feedback.
Q12: “d-VAE itself should also be explained more clearly in the main text. Currently, only the high-level idea of the objective is explained. The explanation should be more precise and include the idea of encoding to latent state, explain the relation to pip-VAE, explain inputs and outputs, linearity/nonlinearity of various mappings, etc. Also see comment 1 above, where I suggest adding more details about other methods in the main text.”
Our primary objective is to delve into the encoding and decoding mechanisms using the separated relevant signals. Therefore, providing an excessive amount of model details could potentially distract from the main focus of the paper. In response to your suggestion, we have included a visual representation of d-VAE's structure, input, and output (see Fig. S1) in the revised manuscript, which offers a comprehensive and intuitive overview. Additionally, we have expanded on the details of d-VAE and other methods in the Methods section.
Thank you for your valuable feedback.
Q13: “In Fig 1f and g, shouldn't the performance plots be swapped? The current plots seem counterintuitive. If there is bias toward decoding (panel g), why is the irrelevant residual so good at decoding?”
The placement of the performance plots in Fig. 1f and 1g is accurate. When the model exhibits a bias toward decoding, it prioritizes extracting the most relevant features (latent variables) for decoding purposes. As a consequence, the model predominantly generates signals that are closely associated with these extracted features. This selective signal extraction and generation process may result in the exclusion of other potentially useful information, which will be left in the residuals. To illustrate this concept, consider the example of face recognition: if a model can accurately identify an individual using only the person's eyes (assuming these are the most useful features), other valuable information, such as details of the nose or mouth, will be left in the residuals, which could also be used to identify the individual.
Thank you for your valuable feedback.
-

eLife assessment
This study presents a useful method for the extraction of behaviour-related activity from neural population recordings based on a specific deep learning architecture - a variational autoencoder. However, the evidence supporting the scientific claims resulting from the application of this method is incomplete as the results may stem, in part, from its properties. The main limitations are: (1) benchmarking against comparable methods is limited; and (2) some observations may be a byproduct of their method, and may not constitute new scientific observations.
-

Reviewer #1 (Public Review):
This work seeks to understand how behaviour-related information is represented in the neural activity of the primate motor cortex. To this end, a statistical model of neural activity is presented that enables a non-linear separation of behaviour-related from unrelated activity. As a generative model, it enables the separate analysis of these two activity modes, here primarily done by assessing the decoding performance of hand movements the monkeys perform in the experiments. Several lines of analysis are presented to show that while the neurons with significant tuning to movements strongly contribute to the behaviourally-relevant activity subspace, less or un-tuned neurons also carry decodable information. It is further shown that the discovered subspaces enable linear decoding, leading the authors to …
Reviewer #1 (Public Review):
This work seeks to understand how behaviour-related information is represented in the neural activity of the primate motor cortex. To this end, a statistical model of neural activity is presented that enables a non-linear separation of behaviour-related from unrelated activity. As a generative model, it enables the separate analysis of these two activity modes, here primarily done by assessing the decoding performance of hand movements the monkeys perform in the experiments. Several lines of analysis are presented to show that while the neurons with significant tuning to movements strongly contribute to the behaviourally-relevant activity subspace, less or un-tuned neurons also carry decodable information. It is further shown that the discovered subspaces enable linear decoding, leading the authors to conclude that motor cortex read-out can be linear.
Strengths:
In my opinion, using an expressive generative model to analyse neural state spaces is an interesting approach to understand neural population coding. While potentially sacrificing interpretability, this approach allows capturing both redundancies and synergies in the code as done in this paper. The model presented here is a natural non-linear extension of a previous linear model PSID) and uses weak supervision in a manner similar to a previous non-linear model (TNDM).
Weaknesses:
This revised version provides additional evidence to support the author's claims regarding model performance and interpretation of the structure of the resulting latent spaces, in particular the distributed neural code over the whole recorded population, not just the well-tuned neurons. The improved ability to linearly decode behaviour from the relevant subspace and the analysis of the linear subspace projections in my opinion convincingly demonstrates that the model picks up behaviour-relevant dynamics, and that these are distributed widely across the population. As reviewer 3 also points out, I would, however, caution to interpret this as evidence for linear read-out of the motor system - your model performs a non-linear transformation, and while this is indeed linearly decodable, the motor system would need to do something similar first to achieve the same. In fact to me it seems to show the opposite, that behaviour-related information may not be generally accessible to linear decoders (including to down-stream brain areas).
As in my initial review, I would also caution against making strong claims about identifiability although this work and TNDM seem to show that in practise such methods work quite well. CEBRA, in contrast, offers some theoretical guarantees, but it is not a generative model, so would not allow the type of analysis done in this paper. In your model there is a para,eter \alpha to balance between neural and behaviour reconstruction. This seems very similar to TNDM and has to be optimised - if this is correct, then there is manual intervention required to identify a good model.
Somewhat related, I also found that the now comprehensive comparison with related models shows that the using decoding performance (R2) as a metric for model comparison may be problematic: the R2 values reported in Figure 2 (e.g. the MC_RTT dataset) should be compared to the values reported in the neural latent benchmark, which represent well-tuned models (e.g. AutoLFADS). The numbers (difficult to see, a table with numbers in the appendix would be useful, see: https://eval.ai/web/challenges/challenge-page/1256/leaderboard) seem lower than what can be obtained with models without latent space disentanglement. While this does not necessarily invalidate the conclusions drawn here, it shows that decoding performance can depend on a variety of model choices, and may not be ideal to discriminate between models. I'm also surprised by the low neural R2 for LFADS I assume this is condition-averaged) - LFADS tends to perform very well on this metric.
One statement I still cannot follow is how the prior of the variational distribution is modelled. You say you depart from the usual Gaussian prior, but equation 7 seems to suggest there is a normal prior. Are the parameters of this distribution learned? As I pointed out earlier, I however suspect this may not matter much as you give the prior a very low weight. I also still am not sure how you generate a sample from the variational distribution, do you just draw one for each pass?
Summary:
This paper presents a very interesting analysis, but some concerns remain that mainly stem from the complexity of deep learning models. It would be good to acknowledge these as readers without relevant background need to understand where the possible caveats are.
-

Reviewer #2 (Public Review):
Li et al present a method to extract "behaviorally relevant" signals from neural activity. The method is meant to solve a problem which likely has high utility for neuroscience researchers. There are numerous existing methods to achieve this goal some of which the authors compare their method to-thankfully, the revised version includes one of the major previous omissions (TNDM). However, I still believe that d-VAE is a promising approach that has its own advantages. Still, I have issues with the paper as-is. The authors have made relatively few modifications to the text based on my previous comments, and the responses have largely just dismissed my feedback and restated claims from the paper. Nearly all of my previous comments remain relevant for this revised manuscript. As such, they have done little to …
Reviewer #2 (Public Review):
Li et al present a method to extract "behaviorally relevant" signals from neural activity. The method is meant to solve a problem which likely has high utility for neuroscience researchers. There are numerous existing methods to achieve this goal some of which the authors compare their method to-thankfully, the revised version includes one of the major previous omissions (TNDM). However, I still believe that d-VAE is a promising approach that has its own advantages. Still, I have issues with the paper as-is. The authors have made relatively few modifications to the text based on my previous comments, and the responses have largely just dismissed my feedback and restated claims from the paper. Nearly all of my previous comments remain relevant for this revised manuscript. As such, they have done little to assuage my concerns, the most important of which I will restate here using the labels/notation (Q1, Q2, etc) from the reviewer response.
Q1) I still remain unconvinced that the core findings of the paper are "unexpected". In the response to my previous Specific Comment #1, they say "We use the term 'unexpected' due to the disparity between our findings and the prior understanding concerning neural encoding and decoding." However, they provide no citations or grounding for why they make those claims. What prior understanding makes it unexpected that encoding is more complex than decoding given the entropy, sparseness, and high dimensionality of neural signals (the "encoding") compared to the smoothness and low dimensionality of typical behavioural signals (the "decoding")?
Q2) I still take issue with the premise that signals in the brain are "irrelevant" simply because they do not correlate with a fixed temporal lag with a particular behavioural feature hand-chosen by the experimenter. In the response to my previous review, the authors say "we employ terms like 'behaviorally-relevant' and 'behaviorally-irrelevant' only regarding behavioral variables of interest measured within a given task, such as arm kinematics during a motor control task.". This is just a restatement of their definition, not a response to my concern, and does not address my concern that the method requires a fixed temporal lag and continual decoding/encoding. My example of reward signals remains. There is a huge body of literature dating back to the 70s on the linear relationships between neural and activity and arm kinematics; in a sense, the authors have chosen the "variable of interest" that proves their point. This all ties back to the previous comment: this is mostly expected, not unexpected, when relating apparently-stochastic, discrete action potential events to smoothly varying limb kinematics.
Q5) The authors seem to have missed the spirit of my critique: to say "linear readout is performed in motor cortex" is an over-interpretation of what their model can show.
Q7) Agreeing with my critique is not sufficient; please provide the data or simulations that provides the context for the reference in the fano factor. I believe my critique is still valid.
Q8) Thank you for comparing to TNDM, it's a useful benchmark.
-

Reviewer #4 (Public Review):
I am a new reviewer for this manuscript, which has been reviewed before. The authors provide a variational autoencoder that has three objectives in the loss: linear reconstruction of behavior from embeddings, reconstruction of neural data, and KL divergence term related to the variational model elements. They take the output of the VAE as the "behaviorally relevant" part of neural data and call the residual "behaviorally irrelevant". Results aim to inspect the linear versus nonlinear behavior decoding using the original raw neural data versus the inferred behaviorally relevant and irrelevant parts of the signal.
Overall, studying neural computations that are behaviorally relevant or not is an important problem, which several previous studies have explored (for example PSID in (Sani et al. 2021), TNDM in …
Reviewer #4 (Public Review):
I am a new reviewer for this manuscript, which has been reviewed before. The authors provide a variational autoencoder that has three objectives in the loss: linear reconstruction of behavior from embeddings, reconstruction of neural data, and KL divergence term related to the variational model elements. They take the output of the VAE as the "behaviorally relevant" part of neural data and call the residual "behaviorally irrelevant". Results aim to inspect the linear versus nonlinear behavior decoding using the original raw neural data versus the inferred behaviorally relevant and irrelevant parts of the signal.
Overall, studying neural computations that are behaviorally relevant or not is an important problem, which several previous studies have explored (for example PSID in (Sani et al. 2021), TNDM in (Hurwitz et al. 2021), TAME-GP in (Balzani et al. 2023), pi-VAE in (Zhou and Wei 2020), and dPCA in (Kobak et al. 2016), etc). However, this manuscript does not properly put their work in the context of such prior works. For example, the abstract states "One solution is to accurately separate behaviorally-relevant and irrelevant signals, but this approach remains elusive", which is not the case given that these prior works have done that. The same is true for various claims in the main text, for example "Furthermore, we found that the dimensionality of primary subspace of raw signals (26, 64, and 45 for datasets A, B, and C) is significantly higher than that of behaviorally-relevant signals (7, 13, and 9), indicating that using raw signals to estimate the neural dimensionality of behaviors leads to an overestimation" (line 321). This finding was presented in (Sani et al. 2021) and (Hurwitz et al. 2021), which is not clarified here. This issue of putting the work in context has been brought up by other reviewers previously but seems to remain largely unaddressed. The introduction is inaccurate also in that it mixes up methods that were designed for separation of behaviorally relevant information with those that are unsupervised and do not aim to do so (e.g., LFADS). The introduction should be significantly revised to explicitly discuss prior models/works that specifically formulated this behavior separation and what these prior studies found, and how this study differs.
Beyond the above, some of the main claims/conclusions made by the manuscript are not properly supported by the analyses and results, which has also been brought up by other reviewers but not fully addressed. First, the analyses here do not support the linear readout from the motor cortex because i) by construction, the VAE here is trained to have a linear readout from its embedding in its loss, which can bias its outputs toward doing well with a linear decoder/readout, and ii) the overall mapping from neural data to behavior includes both the VAE and the linear readout and thus is always nonlinear (even when a linear Kalman filter is used for decoding). This claim is also vague as there is no definition of readout from "motor cortex" or what it means. Why is the readout from the bottleneck of this particular VAE the readout of motor cortex? Second, other claims about properties of individual neurons are also confounded because the VAE is a population-level model that extracts the bottleneck from all neurons. Thus, information can leak from any set of neurons to other sets of neurons during the inference of behaviorally relevant parts of signals. Overall, the results do not convincingly support the claims, and thus the claims should be carefully revised and significantly tempered to avoid misinterpretation by readers.
Below I briefly expand on these as well as other issues, and provide suggestions:
Claims about linearity of "motor cortex" readout are not supported by results yet stated even in the abstract. Instead, what the results support is that for decoding behavior from the output of the dVAE model -- that is trained specifically to have a linear behavior readout from its embedding -- a nonlinear readout does not help. This result can be biased by the very construction of the dVAE's loss that encourages a linear readout/decoding from embeddings, and thus does not imply a finding about motor cortex.
Related to the above, it is unclear what the manuscript means by readout from motor cortex. A clearer definition of "readout" (a mapping from what to what?) in general is needed. The mapping that the linearity/nonlinearity claims refer to is from the *inferred* behaviorally relevant neural signals, which themselves are inferred nonlinearly using the VAE. This should be explicitly clarified in all claims, i.e., that only the mapping from distilled signals to behavior is linear, not the whole mapping from neural data to behavior. Again, to say the readout from motor cortex is linear is not supported, including in the abstract.
Claims about individual neurons are also confounded. The d-VAE distilling processing is a population level embedding so the individual distilled neurons are not obtainable on their own without using the population data. This population level approach also raises the possibility that information can leak from one neuron to another during distillation, which is indeed what the authors hope would recover true information about individual neurons that wasn't there in the recording (the pixel denoising example). The authors acknowledge the possibility that information could leak to a neuron that didn't truly have that information and try to rule it out to some extent with some simulations and by comparing the distilled behaviorally relevant signals to the original neural signals. But ultimately, the distilled signals are different enough from the original signals to substantially improve decoding of low information neurons, and one cannot be sure if all of the information in distilled signals from any individual neuron truly belongs to that neuron. It is still quite likely that some of the improved behavior prediction of the distilled version of low-information neurons is due to leakage of behaviorally relevant information from other neurons, not the former's inherent behavioral information. This should be explicitly acknowledged in the manuscript.
Given the nuances involved in appropriate comparisons across methods and since two of the datasets are public, the authors should provide their complete code (not just the dVAE method code), including the code for data loading, data preprocessing, model fitting and model evaluation for all methods and public datasets. This will alleviate concerns and allow readers to confirm conclusions (e.g., figure 2) for themselves down the line.
Related to 1) above, the authors should explore the results if the affine network h(.) (from embedding to behavior) was replaced with a nonlinear ANN. Perhaps linear decoders would no longer be as close to nonlinear decoders. Regardless, the claim of linearity should be revised as described in 1) and 2) above, and all caveats should be discussed.
The beginning of the section on the "smaller R2 neurons" should clearly define what R2 is being discussed. Based on the response to previous reviewers, this R2 "signifies the proportion of neuronal activity variance explained by the linear encoding model, calculated using raw signals". This should be mentioned and made clear in the main text whenever this R2 is referred to.
Various terms require clear definitions. The authors sometimes use vague terminology (e.g., "useless") without a clear definition. Similarly, discussions regarding dimensionality could benefit from more precise definitions. How is neural dimensionality defined? For example, how is "neural dimensionality of specific behaviors" (line 590) defined? Related to this, I agree with Reviewer 2 that a clear definition of irrelevant should be mentioned that clarifies that relevance is roughly taken as "correlated or predictive with a fixed time lag". The analyses do not explore relevance with arbitrary time lags between neural and behavior data.
CEBRA itself doesn't provide a neural reconstruction from its embeddings, but one could obtain one via a regression from extracted CEBRA embeddings to neural data. In addition to decoding results of CEBRA (figure S3), the neural reconstruction of CEBRA should be computed and CEBRA should be added to Figure 2 to see how the behaviorally relevant and irrelevant signals from CEBRA compare to other methods.
References:
Kobak, Dmitry, Wieland Brendel, Christos Constantinidis, Claudia E Feierstein, Adam Kepecs, Zachary F Mainen, Xue-Lian Qi, Ranulfo Romo, Naoshige Uchida, and Christian K Machens. 2016. "Demixed Principal Component Analysis of Neural Population Data." Edited by Mark CW van Rossum. eLife 5 (April): e10989. https://doi.org/10.7554/eLife.10989.
Sani, Omid G., Hamidreza Abbaspourazad, Yan T. Wong, Bijan Pesaran, and Maryam M. Shanechi. 2021. "Modeling Behaviorally Relevant Neural Dynamics Enabled by Preferential Subspace Identification." Nature Neuroscience 24 (1): 140-49. https://doi.org/10.1038/s41593-020-00733-0.
Zhou, Ding, and Xue-Xin Wei. 2020. "Learning Identifiable and Interpretable Latent Models of High-Dimensional Neural Activity Using Pi-VAE." In Advances in Neural Information Processing Systems, 33:7234-47. Curran Associates, Inc. https://proceedings.neurips.cc/paper/2020/hash/510f2318f324cf07fce24c3a4b89c771-Abstract.html.
Hurwitz, Cole, Akash Srivastava, Kai Xu, Justin Jude, Matthew Perich, Lee Miller, and Matthias Hennig. 2021. "Targeted Neural Dynamical Modeling." In Advances in Neural Information Processing Systems. Vol. 34. https://proceedings.neurips.cc/paper/2021/hash/f5cfbc876972bd0d031c8abc37344c28-Abstract.html.
Balzani, Edoardo, Jean-Paul G. Noel, Pedro Herrero-Vidal, Dora E. Angelaki, and Cristina Savin. 2023. "A Probabilistic Framework for Task-Aligned Intra- and Inter-Area Neural Manifold Estimation." In . https://openreview.net/forum?id=kt-dcBQcSA.
-

eLife assessment
This study presents a useful method for the extraction of behaviour-related activity from neural population recordings based on a specific deep learning architecture - a variational autoencoder. However, the evidence supporting the scientific claims resulting from the application of this method is incomplete as the results may stem, in part, from its properties. The authors should: (1) improve how they benchmark their method, by comparing against additional relevant techniques, and (2) reframe their results considering what observations may be a byproduct of their method, and which do constitute new scientific observations.
-

Reviewer #1 (Public Review):
This work seeks to understand how behaviour-related information is represented in the neural activity of the primate motor cortex. To this end, a statistical model of neural activity is presented that enables a non-linear separation of behaviour-related from unrelated activity. As a generative model, it enables the separate analysis of these two activity modes, here primarily done by assessing the decoding performance of hand movements the monkeys perform in the experiments. Several lines of analysis are presented to show that while the neurons with significant tuning to movements strongly contribute to the behaviourally-relevant activity subspace, less or un-tuned neurons also carry decodable information. It is further shown that the discovered subspaces enable linear decoding, leading the authors to …
Reviewer #1 (Public Review):
This work seeks to understand how behaviour-related information is represented in the neural activity of the primate motor cortex. To this end, a statistical model of neural activity is presented that enables a non-linear separation of behaviour-related from unrelated activity. As a generative model, it enables the separate analysis of these two activity modes, here primarily done by assessing the decoding performance of hand movements the monkeys perform in the experiments. Several lines of analysis are presented to show that while the neurons with significant tuning to movements strongly contribute to the behaviourally-relevant activity subspace, less or un-tuned neurons also carry decodable information. It is further shown that the discovered subspaces enable linear decoding, leading the authors to conclude that motor cortex read-out can be linear.
Strengths:
In my opinion, using an expressive generative model to analyse neural state spaces is an interesting approach to understanding neural population coding. While potentially sacrificing interpretability, this approach allows capturing both redundancies and synergies in the code as done in this paper. The model presented here is a natural non-linear extension of a previous linear model (PSID) and
Weaknesses:
First, the model in the paper is almost identical to an existing VAE model (TNDM) that makes use of weak supervision with behaviour in the same way [1]. This paper should at least be referenced. If the authors wish they could compare their model to TNDM, which combines a state space model with smoothing similar to LFADS. Given that TNDM achieves very good behaviour reconstructions, it may be on par with this model without the need for a Kalman filter (and hence may achieve better separation of behaviour-related and unrelated dynamics).
Second, in my opinion, the claims regarding identifiability are overstated - this matters as the results depend on this to some extent. Recent work shows that VAEs generally suffer from identifiability problems due to the Gaussian latent space [2]. This paper also hints that weak supervision may help to resolve such issues, so this model as well as TNDM and CEBRA may indeed benefit from this. In addition however, it appears that the relative weight of the KL Divergence in the VAE objective is chosen very small compared to the likelihood (0.1%), so the influence of the prior is weak and the model may essentially learn the average neural trajectories while underestimating the noise in the latent variables. This, in turn, could mean that the model will not autoencode neural activity as well as it should, note that an average R2 in this case will still be high (I could not see how this is actually computed). At the same time, the behaviour R2 will be large simply because the different movement trajectories are very distinct. Since the paper makes claims about the roles of different neurons, it would be important to understand how well their single trial activities are reconstructed, which can perhaps best be investigated by comparing the Poisson likelihood (LFADS is a good baseline model). Taken together, while it certainly makes sense that well-tuned neurons contribute more to behaviour decoding, I worry that the very interesting claim that neurons with weak tuning contain behavioural signals is not well supported.
Third, and relating to this issue, I could not entirely follow the reasoning in the section arguing that behavioural information can be inferred from neurons with weak selectivity, but that it is not linearly decodable. It is right to test if weak supervision signals bleed into the irrelevant subspace, but I could not follow the explanations. Why, for instance, is the ANN decoder on raw data (I assume this is a decoder trained fully supervised) not equal in performance to the revenant distilled signals? Should a well-trained non-linear decoder not simply yield a performance ceiling? Next, if I understand correctly, distilled signals were obtained from the full model. How does a model perform trained only on the weakly tuned neurons? Is it possible that the subspaces obtained with the model are just not optimally aligned for decoding? This could be a result of limited identifiability or model specifics that bias reconstruction to averages (a well-known problem of VAEs). I, therefore, think this analysis should be complemented with tests that do not depend on the model.
Finally, a more technical issue to note is related to the choice to learn a non-parametric prior instead of using a conventional Gaussian prior. How is this implemented? Is just a single sample taken during a forward pass? I worry this may be insufficient as this would not sample the prior well, and some other strategy such as importance sampling may be required (unless the prior is not relevant as it weakly contributed to the ELBO, in which case this choice seems not very relevant). Generally, it would be useful to see visualisations of the latent variables to see how information about behaviour is represented by the model.
Summary:
This paper presents a very interesting analysis, but I have several concerns as to well the analysis supports the main conclusions. I think the work could benefit from an additional complementary analysis that seeks to confirm with another method if weakly tuned neurons indeed show an encoding that differs qualitatively from the strongly tuned ones.
[1] Hurwitz, Cole, et al. "Targeted neural dynamical modeling." Advances in Neural Information Processing Systems 34 (2021): 29379-29392.
[2] Hyvarinen, Aapo, Ilyes Khemakhem, and Hiroshi Morioka. "Nonlinear Independent Component Analysis for Principled Disentanglement in Unsupervised Deep Learning." arXiv preprint arXiv:2303.16535 (2023). -

Reviewer #2 (Public Review):
Li et al present a method to extract "behaviorally relevant" signals from neural activity. The method is meant to solve a problem which likely has high utility for neuroscience researchers. There are numerous existing methods to achieve this goal some of which the authors compare their method to, though there are notable omissions. However, I do believe that d-VAE is a promising approach that has its own advantages.
That being said, there are issues with the paper as-is. This could have been a straightforward "methods" paper describing their approach and validating it on different ground truth and experimental datasets. Instead, the authors focus on the neuroscientific results and their implications for brain mechanisms. Unfortunately, while the underlying method seems sound and performs well relative to the …
Reviewer #2 (Public Review):
Li et al present a method to extract "behaviorally relevant" signals from neural activity. The method is meant to solve a problem which likely has high utility for neuroscience researchers. There are numerous existing methods to achieve this goal some of which the authors compare their method to, though there are notable omissions. However, I do believe that d-VAE is a promising approach that has its own advantages.
That being said, there are issues with the paper as-is. This could have been a straightforward "methods" paper describing their approach and validating it on different ground truth and experimental datasets. Instead, the authors focus on the neuroscientific results and their implications for brain mechanisms. Unfortunately, while the underlying method seems sound and performs well relative to the assessed competition, the scientific results and presentation they put forward were not sufficiently strong to support these claims, especially given the small amount of data (recordings of one monkey per task, with considerable variability between them).
Specific comments
- Is the apparently increased complexity of encoding vs decoding so unexpected given the entropy, sparseness, and high dimensionality of neural signals (the "encoding") compared to the smoothness and low dimensionality of typical behavioural signals (the "decoding") recorded in neuroscience experiments? This is the title of the paper so it seems to be the main result on which the authors expect readers to focus.- I take issue with the premise that signals in the brain are "irrelevant" simply because they do not correlate with a fixed temporal lag with a particular behavioural feature hand-chosen by the experimenter. As an example, the presence of a reward signal in motor cortex [1] after the movement is likely to be of little use from the perspective of predicting kinematics from time-bin to time-bin using a fixed model across trials (the apparent definition of "relevant" for behaviour here), but an entire sub-field of neuroscience is dedicated to understanding the impact of these reward-related signals on future behaviour. Is there method sophisticated enough to see the behavioural "relevance" of this brief, transient, post-movement signal? This may just be an issue of semantics, and perhaps I read too much into the choice of words here. Perhaps the authors truly treat "irrelevant" and "without a fixed temporal correlation" as synonymous phrases and the issue is easily resolved with a clarifying parenthetical the first time the word "irrelevant" is used. But I remain troubled by some claims in the paper which lead me to believe that they read more deeply into the "irrelevancy" of these components.
- The authors claim the "irrelevant" responses underpin an unprecedented neuronal redundancy and reveal that movement behaviors are distributed in a higher-dimensional neural space than previously thought." Perhaps I just missed the logic, but I fail to see the evidence for this. The neural space is a fixed dimensionality based on the number of neurons. A more sparse and nonlinear distribution across this set of neurons may mean that linear methods such as PCA are not effective ways to approximate the dimensionality. But ultimately the behaviourally relevant signals seem quite low-dimensional in this paper even if they show some nonlinearity may help.
- Relatedly, I would like to note that the exercise of arbitrarily dividing a continuous distribution of a statistic (the "R2") based on an arbitrary threshold is a conceptually flawed exercise. The authors read too much into the fact that neurons which have a low R2 w.r.t. PDs have behavioural information w.r.t. other methods. To this reviewer, it speaks more about the irrelevance, so to speak, of the preferred direction metric than anything fundamental about the brain.
- there is an apparent logical fallacy that begins in the abstract and persists in the paper: "Surprisingly, when incorporating often-ignored neural dimensions, behavioral information can be decoded linearly as accurately as nonlinear decoding, suggesting linear readout is performed in motor cortex." Don't get me wrong: the equivalency of linear and nonlinear decoding approaches on this dataset is interesting, and useful for neuroscientists in a practical sense. However, the paper expends much effort trying to make fundamental scientific claims that do not feel very strongly supported. This reviewer fails to see what we can learn about a set of neurons in the brain which are presumed to "read out" from motor cortex. These neurons will not have access to the data analyzed here. That a linear model can be conceived by an experimenter does not imply that the brain must use a linear model. The claim may be true, and it may well be that a linear readout is implemented in the brain. Other work [2,3] has shown that linear readouts of nonlinear neural activity patterns can explain some behavioural features. The claim in this paper, however, is not given enough
- I am afraid I may be missing something, as I did not understand the fano factor analysis of Figure 3. In a sense the behaviourally relevant signals must have lower FF given they are in effect tied to the temporally smooth (and consistent on average across trials) behavioural covariates. The point of the original Churchland paper was to show that producing a behaviour squelches the variance; naturally these must appear in the behaviourally relevant components. A control distribution or reference of some type would possibly help here.
- The authors compare the method to LFADS. While this is a reasonable benchmark as a prominent method in the field, LFADS does not attempt to solve the same problem as d-VAE. A better and much more fair comparison would be TNDM [4], an extension of LFADS which is designed to identify behaviourally relevant dimensions.
[1] https://doi.org/10.1371/journal.pone.0160851
[2] https://doi.org/10.1101/2022.03.31.486635
[3] https://doi.org/10.1038/s41593-017-0028-6
[4] Hurwitz et al, Targeted Neural Dynamical Modeling, NeurIPS 2021. -

Reviewer #3 (Public Review):
The authors develop a variational autoencoder (VAE), termed d-VAE (or distill VAE) that aims to tease apart the behaviorally relevant and irrelevant sections of each neuron's firing rate. The input to the VAE is the population activity for a given time step, and the output is the inferred behaviorally relevant section of the population activity at that time step. The residual is referred to as behaviorally irrelevant: total neural activity = behaviorally relevant + behaviorally irrelevant (x = x_r + x_i). The mapping from the raw neural signals (x) to the bottlenecked latent in the autoencoder (called z, z=f(x)) and back to the inferred behaviorally relevant single-neuron activities (x_r = g(z)) is applied per time step (does not incorporate any info from past/future time steps) and, critically, it is …
Reviewer #3 (Public Review):
The authors develop a variational autoencoder (VAE), termed d-VAE (or distill VAE) that aims to tease apart the behaviorally relevant and irrelevant sections of each neuron's firing rate. The input to the VAE is the population activity for a given time step, and the output is the inferred behaviorally relevant section of the population activity at that time step. The residual is referred to as behaviorally irrelevant: total neural activity = behaviorally relevant + behaviorally irrelevant (x = x_r + x_i). The mapping from the raw neural signals (x) to the bottlenecked latent in the autoencoder (called z, z=f(x)) and back to the inferred behaviorally relevant single-neuron activities (x_r = g(z)) is applied per time step (does not incorporate any info from past/future time steps) and, critically, it is nonlinear (f and g are nonlinear feedforward neural networks). The key technical novelty that encourages x_r to encode behaviorally relevant information is a term added to the loss, which penalizes bad linear behavior decoding from the latent z. Otherwise the method is very similar to a prior method called pi-VAE, which should be explained more thoroughly in the manuscript to clearly highlight the technical novelty.
The authors apply their method to 3 non-human primate datasets to infer behaviorally relevant signals and contrast them with the raw neural signals and the residual behaviorally irrelevant signals. As a key performance metric, they compute the accuracy of decoding behavior from the inferred behaviorally relevant signals (x_r) using a linear Kalman filter (KF) or alternatively using a nonlinear feed forward neural network (ANN). They highlight 3 main conclusions in the abstract: first, that single neurons from which behavior is very poorly decodable do encode considerable behavior information in a nonlinear manner, which the ANN can decode. Second, they conclude from various analyses that behavior is occupying a higher dimensional neural space than previously thought. Third, they find that linear KF decoding and nonlinear ANN decoding perform similarly when provided with the inferred behaviorally relevant signals (x_r), from which they conclude that a linear readout must be performed in motor cortex.
The paper is well-written in many places and has high-quality graphics. The questions that it aims to address are also of considerable interest in neuroscience. However, unfortunately, several main conclusions, including but not limited to all 3 conclusions that are highlighted in the abstract, are not fully supported by the results due to confounds, some of which are fundamental to the method. Several statements in the text also seem inaccurate due to use of imprecise language. Moreover, the authors fail to compare with some more relevant existing methods that are specifically designed for extracting behaviorally relevant signals. In addition, for some of the methods they compare with, they do not use an appropriate setup for the benchmark methods, rendering the validation of the proposed method unconvincing. Finally, in many places imprecise language that is not accompanied with an operational definition (e.g., smaller R2 [of what], similar [per what metric]) makes results hard to follow, unless most of the text is read very carefully. Some key details of the methods are also not explained anywhere.
-

-

-

-

-

-