Transcription factors underlying photoreceptor diversity
Curation statements for this article:-
Curated by eLife
Evaluation Summary:
This manuscript offers a valuable transcriptomic data set of known types of adult zebrafish photoreceptors (rod and cones). The study further identifies a large set of differentially expressed transcription factors, many of which still have an unidentified function in photoreceptors. Using CRISPR F0 screening, the study shows that the two tbx2 zebrafish paralogues are involved in photoreceptors specification beyond what is currently known. The study uses a solid methodology and the results will be valuable for researchers interested in photoreceptor biology. At present, however, the manuscript has a misleading title and focus: the analysis of adult photoreceptors can hardly offer a scenario of the transcription factors involved in the specification of photoreceptors.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #3 agreed to share their name with the authors.)
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
During development, retinal progenitors navigate a complex landscape of fate decisions to generate the major cell classes necessary for proper vision. Transcriptional regulation is critical to generate diversity within these major cell classes. Here, we aim to provide the resources and techniques required to identify transcription factors necessary to generate and maintain diversity in photoreceptor subtypes, which are critical for vision. First, we generate a key resource: a high-quality and deep transcriptomic profile of each photoreceptor subtype in adult zebrafish. We make this resource openly accessible, easy to explore, and have integrated it with other currently available photoreceptor transcriptomic datasets. Second, using our transcriptomic profiles, we derive an in-depth map of expression of transcription factors in photoreceptors. Third, we use efficient CRISPR-Cas9 based mutagenesis to screen for null phenotypes in F0 larvae (F0 screening) as a fast, efficient, and versatile technique to assess the involvement of candidate transcription factors in the generation of photoreceptor subtypes. We first show that known phenotypes can be easily replicated using this method: loss of S cones in foxq2 mutants and loss of rods in nr2e3 mutants. We then identify novel functions for the transcription factor Tbx2, demonstrating that it plays distinct roles in controlling the generation of all photoreceptor subtypes within the retina. Our study provides a roadmap to discover additional factors involved in this process. Additionally, we explore four transcription factors of unknown function (Skor1a, Sall1a, Lrrfip1a, and Xbp1), and find no evidence for their involvement in the generation of photoreceptor subtypes. This dataset and screening method will be a valuable way to explore the genes involved in many other essential aspects of photoreceptor biology.
Article activity feed
-

-

Author Response
Reviewer #2 (Public Review):
This paper by Angueyra, et al., adds to the field’s current understanding of photoreceptor specification and factors regulating opsin expression in vertebrates. Current models of specification of vertebrate photoreceptors are largely based on studies of mammals. However, a great number of animals including teleosts express a wider array of photoreceptor subtypes. Zebrafish for example have 4 distinct cone subtypes and rods. The approach is sound and the data are quite convincing. The only minor weaknesses are that the statistical analyses need to be revisited and the discussion should be a bit more focused.
To identify differentially expressed transcription factors, the authors performed bulk RNA-seq of pooled, hand-sorted photoreceptors. The selection criterion was tightly controlled to …
Author Response
Reviewer #2 (Public Review):
This paper by Angueyra, et al., adds to the field’s current understanding of photoreceptor specification and factors regulating opsin expression in vertebrates. Current models of specification of vertebrate photoreceptors are largely based on studies of mammals. However, a great number of animals including teleosts express a wider array of photoreceptor subtypes. Zebrafish for example have 4 distinct cone subtypes and rods. The approach is sound and the data are quite convincing. The only minor weaknesses are that the statistical analyses need to be revisited and the discussion should be a bit more focused.
To identify differentially expressed transcription factors, the authors performed bulk RNA-seq of pooled, hand-sorted photoreceptors. The selection criterion was tightly controlled to limit unhealthy cells and cellular debris from other photoreceptors subtypes. The pooling of cells provided a considerable depth of sequencing, orders of magnitude better than scSeq. The authors identified known transcription factors and several that appear to be novel or their role has not been determined. The data are made available on the PIs website as is a program to access and compare the gene expression data.
The authors then used CRISPR/Cas9 gene targeting of two known and several novel factors identified in their analysis for effects on cell fate decisions and opsin expression. Phenotyping performed on the injected larvae is possible, and the target genes were applied and sequenced to demonstrate the efficiency of the gene targeting. Targeting of 2 genes with know functions in photoreceptor specification in zebrafish, Tbx2b and Foxq2 resulted in the anticipated changes in cell fate, albeit, the strength of the alterations in cell fate in the F0 larvae appears to be less than the published phenotypes for the inherited alleles. Interestingly, the authors also identified the expression of an RH2 opsin in the SWS2 another cone type. The changes are subtle but important.
The authors then targeted tbx2a, the function of which was not known. The result is quite interesting as it matches the increase of rods and decrease of UV cones observed in tbx2b mutants. However, the injected animals also showed RH2 opsin expression but are now in the LWS cone subtype. These data suggest that Tbx2 transcription factors repress misexpression of opsins in the wrong cell type.
The authors also show that targeting additional differentially expressed factors does not affect photoreceptor fate or survival in the time frame investigated. These are important data to present. For these or any of the other targeted genes above, did the authors test for changes in photoreceptor number or survival?
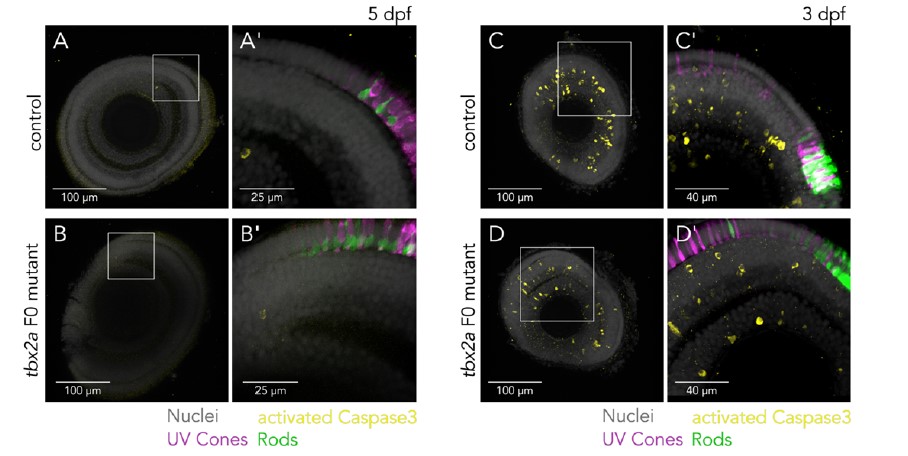
We have attempted to address this point, but the answer is not clear cut. We used activated caspase-3 inmmunolabeling as a marker of apoptosis (Lusk and Kwan 2022). At 5 dpf, the age we chose to make quantifications, we don’t see an increase in activated caspase-3 positive cells when we compare control and tbx2a F0 mutants (Reviewer Figure 1A-B). Labeled cells are very rare and located near the ciliary marginal zone irrespective of genotype. This suggests that there is no detectable active death at this late stage of development in tbx2 F0 mutants. Earlier in development, at 3 dpf, when photoreceptor subtypes first appear, there is also a normal wave of apoptosis in the retina (Blume et al. 2020; Biehlmaier, Neuhauss, and Kohler 2001), resulting in many cells positive for activated caspase-3; our preliminary quantifications don’t show a marked increase in the number of labeled cells in tbx2a F0 mutants, but we consider that it’s likely that subtle effects might be obscured by the physiological wave of apoptosis (Reviewer Figure 1C-D).
Reviewer Figure 1 - Assessment of apoptosis in tbx2a F0 mutants. (A-B) Confocal images of 5 dpf larval eyes of control (A and A’) and tbx2a F0 mutants (B and B’) counterstained with DAPI (grey) and immunolabeled against activated Caspase 3 (yellow) show sparse and dim labeling, restricted to cells located in the ciliary marginal zone, without clear differences between groups. (C-D) Confocal images of 3 dpf larval eyes of control (C and C’) and tbx2a F0 mutants (D and D’) immunolabeled against activated Caspase 3 show many positive cells, located in all retinal layers, as expected from physiological apoptosis at this stage of development and without clear differences between groups.
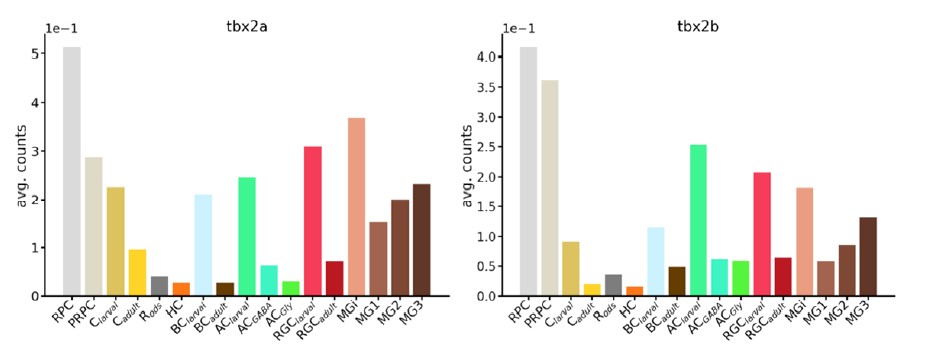
Furthermore, the additional single-cell RNA-seq datasets we have reanalyzed suggest that tbx2a and tbx2b are expressed by other retinal neurons and progenitors and not just photoreceptors (Reviewer Figure 2), further confounding attempts at the quantification of apoptosis specifically in photoreceptor progenitors.
Reviewer Figure 2 – Expression of tbx2 paralogues across retinal cell types. The transcription factors tbx2a and tbx2b are expressed by many retinal cells. Plots show average counts across clusters in RNA-seq data obtained by Hoang et al. (2020).
At this stage, we consider that fully resolving this issue is important and will require considerably more work, which we will pursue in the future using full germline mutants and live-imaging experiments.
Reviewer #3 (Public Review):
Angueyra et al. tried to establish the method to identify key factors regulating fate decisions in the retinal visual photoreceptor cells by combining transcriptomic and fast genome editing approaches. First, they isolated and pooled five subtypes of photoreceptor cells from the transgenic lines in each of which a specific subtype of photoreceptor cells are labeled by fluorescence protein, and then subjected them to RNA-seq analyses. Second, by comparing the transcriptome data, they extracted the list of the transcription factor genes enriched in the pooled samples. Third, they applied CRISPR-based F0 knockout to functionally identify transcription factor genes involved in cell fate decisions of photoreceptor subtypes. To benchmark this approach, they initially targeted foxq2 and nr2e3 genes, which have been previously shown to regulate S-opsin expression and S-cone cell fate (foxq2) and to regulate rhodopsin expression and rod fate (nr2e3). They then targeted other transcription factor genes in the candidate list and found that tbx2a and tbx2b are independently required for UV-cone specification. They also found that tbx2a expressed in the L-cone subtype and tbx2b expressed in L-cones inhibit M-opsin gene expression in the respective cone subtypes. From these data, the authors concluded that the transcription factors Tbx2a and Tbx2b play a central role in controlling the identity of all photoreceptor subtypes within the retina.
Overall, the contents of this manuscript are well organized and technically sound. The authors presented convincing data, and carefully analyzed and interpreted them. It includes an evaluation of the presented data on cell-type specific transcriptome by comparing it with previously published ones. I think the current transcriptomic data will be a valuable platform to identify the genes regulating cell-type specific functions, especially in combination with the fast CRISPR-based in vivo screening methods provided here. I hope that the following points would be helpful for the authors to improve the manuscript appropriately.
- The manuscript uses the word “FØ” quite often without any proper definition. I wonder how “Ø” should be pronounced - zero or phi? This word is not common and has not been used in previous publications. I feel the phrase “F0 knockout,” which was used in the paper cited by the authors (Kroll et al 2021), is more straightforward. If it is to be used in the manuscript, please define “FØ” and “CRISPR-FØ screening” appropriately, especially in the abstract.
We have made changes to replace “FØ” to “F0.” In our other citation (Hoshijima et al., 2019), “F0 embryo” was used throughout the paper. Following our references and Dr Kojima’s suggestion, we adopted “F0 mutant larva” as the most straightforward and less confusing term. We have also made changes in the abstract to define our approach more clearly and made appropriate changes throughout the manuscript.
- Figure 1-supplement 1 shows that opn1mw4 has quite high (normalized) FPKM in one of the S-cone samples in contrast to the least (or no) expression in the M-cone samples, in which opn1mw4 is expected to be detected. The authors should address a possible origin of this inconsistent result for opn1mw4 expression as well as a technical limitation of using the Tg(opn1mw2:egfp) line for detection of opn1mw4 expression in the GFP-positive cells.
In Figure 1 - Supplement 1, we had attempted to provide a summarized figure of all phototransduction genes, but the big differences in expression levels — in particular, the high expression of opsins genes — forced us to use gene-by-gene normalization for display. Without normalization, the expression of opn1mw4 is very low across all samples, and its detection in that sole S-cone sample can likely be attributed to some degree of inherent noise in our methods. We have revised Figure 1 - Supplement 1: we find that we can avoid gene-by-gene normalization and still provide a good summary of the expression of phototransduction genes if the heatmap is broken down by gene families, which have more similar expression levels. In addition, we have added caveats to the use of the Tg(opn1mw2:egfp) line as our sole M-cone marker in the results section describing our RNA-seq approach, including our inability to provide data on Opn1mw4-expressing M cones.
- The manuscript lacks a description of the sampling time point. It is well known that many genes are expressed with daily (or circadian) fluctuation (cf. Doherty & Kay, 2010 Annu. Rev. Genet.). For example, the cone-specific gene list in Fig.2C includes a circadian clock gene, per3, whose expression was reported to fluctuate in a circadian manner in many tissues of zebrafish including the retina (Kaneko et al. 2006 PNAS). It appears to be cone-specific at this time point of sample collection as shown in Fig.2, but might be expressed in a different pattern at other time points (eg, rod expression). The authors should add, at least, a clear description of the sampling time points so as to make their data more informative.
We have included this information in the materials and methods. We collected all our samples during the most active peak of the zebrafish circadian rhythm between 11am and 2pm (3h to 6h after light onset) to avoid the influence of circadian fluctuations in our analysis.
-

Evaluation Summary:
This manuscript offers a valuable transcriptomic data set of known types of adult zebrafish photoreceptors (rod and cones). The study further identifies a large set of differentially expressed transcription factors, many of which still have an unidentified function in photoreceptors. Using CRISPR F0 screening, the study shows that the two tbx2 zebrafish paralogues are involved in photoreceptors specification beyond what is currently known. The study uses a solid methodology and the results will be valuable for researchers interested in photoreceptor biology. At present, however, the manuscript has a misleading title and focus: the analysis of adult photoreceptors can hardly offer a scenario of the transcription factors involved in the specification of photoreceptors.
(This preprint has been reviewed by eLife. We …
Evaluation Summary:
This manuscript offers a valuable transcriptomic data set of known types of adult zebrafish photoreceptors (rod and cones). The study further identifies a large set of differentially expressed transcription factors, many of which still have an unidentified function in photoreceptors. Using CRISPR F0 screening, the study shows that the two tbx2 zebrafish paralogues are involved in photoreceptors specification beyond what is currently known. The study uses a solid methodology and the results will be valuable for researchers interested in photoreceptor biology. At present, however, the manuscript has a misleading title and focus: the analysis of adult photoreceptors can hardly offer a scenario of the transcription factors involved in the specification of photoreceptors.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #3 agreed to share their name with the authors.)
-

Reviewer #1 (Public Review):
The present manuscript offers valuable transcriptomic data sets of manually picked adult zebrafish photoreceptors from dissociated retinas of different transgenic lines, in which rods and cones (UV, S, L, M) were marked by the fluorescent reporter proteins. This is a very valuable approach because allows for selecting "healthy cells". Whether the approach is comparable to single-cell RNA-seq as the authors do (see page 3 and discussion) is however questionable as each of their samples is composed of 20 cells.
The authors further focused on transcription factors that are differentially expressed in the five photoreceptors cell types that they analyze, identifying a large number of them with still unidentified functions. This is very valuable information. However, the idea that this analysis will help to …
Reviewer #1 (Public Review):
The present manuscript offers valuable transcriptomic data sets of manually picked adult zebrafish photoreceptors from dissociated retinas of different transgenic lines, in which rods and cones (UV, S, L, M) were marked by the fluorescent reporter proteins. This is a very valuable approach because allows for selecting "healthy cells". Whether the approach is comparable to single-cell RNA-seq as the authors do (see page 3 and discussion) is however questionable as each of their samples is composed of 20 cells.
The authors further focused on transcription factors that are differentially expressed in the five photoreceptors cell types that they analyze, identifying a large number of them with still unidentified functions. This is very valuable information. However, the idea that this analysis will help to identify new TF involved in the specification of the photoreceptors (as stressed in the title) is at odds with the experimental setup. The authors have analyzed adult photoreceptors and thus by definition cells that had been already specified. Many of the TF involved in the specification may no longer be expressed. The analysis rather offers a list of TFs that are involved in photoreceptor homeostasis, some of which had been also involved in their specification. Proof of this is the fact that none of the four TFs of yet uncharacterized function (Skor1a, Sall1a, Lrrfip1a, and Xbp1) turned out to be involved in photoreceptor specification. The F0 screen only confirmed factors that were already known to be involved in cell specification and that in adult photoreceptors likely play a different role.
The authors further investigate the activity of the two tbx2 zebrafish paralogues in photoreceptors' specification, showing a novel role for tbx2 in the repression of different opsin in specific photoreceptor cell types. This is an interesting finding, however, it is overinterpreted by the authors. Indeed, tbx2 cannot be considered as a "master regulator of photoreceptor fate" (page 7) but, at best, a TF is required to control an appropriate proportion of the different photoreceptors' subtypes.
Overall this is an interesting and well-performed study with valuable information. The conceptual framework of the study should however be re-elaborated, further avoiding overinterpretations.
-

Reviewer #2 (Public Review):
This paper by Angueyra, et al., adds to the field's current understanding of photoreceptor specification and factors regulating opsin expression in vertebrates. Current models of specification of vertebrate photoreceptors are largely based on studies of mammals. However, a great number of animals including teleosts express a wider array of photoreceptor subtypes. Zebrafish for example have 4 distinct cone subtypes and rods. The approach is sound and the data are quite convincing. The only minor weaknesses are that the statistical analyses need to be revisited and the discussion should be a bit more focused.
To identify differentially expressed transcription factors, the authors performed bulk RNA-seq of pooled, hand-sorted photoreceptors. The selection criterion was tightly controlled to limit unhealthy …
Reviewer #2 (Public Review):
This paper by Angueyra, et al., adds to the field's current understanding of photoreceptor specification and factors regulating opsin expression in vertebrates. Current models of specification of vertebrate photoreceptors are largely based on studies of mammals. However, a great number of animals including teleosts express a wider array of photoreceptor subtypes. Zebrafish for example have 4 distinct cone subtypes and rods. The approach is sound and the data are quite convincing. The only minor weaknesses are that the statistical analyses need to be revisited and the discussion should be a bit more focused.
To identify differentially expressed transcription factors, the authors performed bulk RNA-seq of pooled, hand-sorted photoreceptors. The selection criterion was tightly controlled to limit unhealthy cells and cellular debris from other photoreceptors subtypes. The pooling of cells provided a considerable depth of sequencing, orders of magnitude better than scSeq. The authors identified known transcription factors and several that appear to be novel or their role has not been determined. The data are made available on the PIs website as is a program to access and compare the gene expression data.
The authors then used Crispr/Cas9 gene targeting of two known and several novel factors identified in their analysis for effects on cell fate decisions and opsin expression. Phenotyping performed on the injected larvae is possible, and the target genes were applied and sequenced to demonstrate the efficiency of the gene targeting. Targeting of 2 genes with know functions in photoreceptor specification in zebrafish, Tbx2b and Foxaq1 resulted in the anticipated changes in cell fate, albeit, the strength of the alterations in cell fate in the F0 larvae appears to be less than the published phenotypes for the inherited alleles. Interestingly, the authors also identified the expression of an RH2 opsin in the SWS2 another cone type. The changes are subtle but important.
The authors then targeted tbx2a, the function of which was not known. The result is quite interesting as it matches the increase of rods and decrease of UV cones observed in tbx2b mutants. However, the injected animals also showed RH2 opsin expression but are now in the LWS cone subtype. These data suggest that Tbx2 transcription factors repress misexpression of opsins in the wrong cell type.
The authors also show that targeting additional differentially expressed factors does not affect photoreceptor fate or survival in the time frame investigated. These are important data to present. For these or any of the other targeted genes above, did the authors test for changes in photoreceptor number or survival?
-

Reviewer #3 (Public Review):
Angueyra et al. tried to establish the method to identify key factors regulating fate decisions in the retinal visual photoreceptor cells by combining transcriptomic and fast genome editing approaches. First, they isolated and pooled five subtypes of photoreceptor cells from the transgenic lines in each of which a specific subtype of photoreceptor cells are labeled by fluorescence protein, and then subjected them to RNAseq analyses. Second, by comparing the transcriptome data, they extracted the list of the transcription factor genes enriched in the pooled samples. Third, they applied CRISPR-based F0 knockout to functionally identify transcription factor genes involved in cell fate decisions of photoreceptor subtypes. To benchmark this approach, they initially targeted foxq2 and nr2e3 genes, which have been …
Reviewer #3 (Public Review):
Angueyra et al. tried to establish the method to identify key factors regulating fate decisions in the retinal visual photoreceptor cells by combining transcriptomic and fast genome editing approaches. First, they isolated and pooled five subtypes of photoreceptor cells from the transgenic lines in each of which a specific subtype of photoreceptor cells are labeled by fluorescence protein, and then subjected them to RNAseq analyses. Second, by comparing the transcriptome data, they extracted the list of the transcription factor genes enriched in the pooled samples. Third, they applied CRISPR-based F0 knockout to functionally identify transcription factor genes involved in cell fate decisions of photoreceptor subtypes. To benchmark this approach, they initially targeted foxq2 and nr2e3 genes, which have been previously shown to regulate S-opsin expression and S-cone cell fate (foxq2) and to regulate rhodopsin expression and rod fate (nr2e3). They then targeted other transcription factor genes in the candidate list and found that tbx2a and tbx2b are independently required for UV-cone specification. They also found that tbx2a expressed in the L-cone subtype and tbx2b expressed in L-cones inhibit M-opsin gene expression in the respective cone subtypes. From these data, the authors concluded that the transcription factors Tbx2a and Tbx2b play a central role in controlling the identity of all photoreceptor subtypes within the retina.
Overall, the contents of this manuscript are well organized and technically sound. The authors presented convincing data, and carefully analyzed and interpreted them. It includes an evaluation of the presented data on cell-type specific transcriptome by comparing it with previously published ones. I think the current transcriptomic data will be a valuable platform to identify the genes regulating cell-type specific functions, especially in combination with the fast CRISPR-based in vivo screening methods provided here. I hope that the following points would be helpful for the authors to improve the manuscript appropriately.
The manuscript uses the word "FØ" quite often without any proper definition. I wonder how "Ø" should be pronounced - zero or phi? This word is not common and has not been used in previous publications. I feel the phrase "F0 knockout", which was used in the paper cited by the authors (Kroll et al 2021), is more straightforward. If it is to be used in the manuscript, please define "FØ" and "CRISPR-FØ screening" appropriately, especially in the abstract.
Figure 1-supplement 1 shows that opn1mw4 has quite high (normalized) FPKM in one of the S-cone samples in contrast to the least (or no) expression in the M-cone samples, in which opn1mw4 is expected to be detected. The authors should address a possible origin of this inconsistent result for opn1mw4 expression as well as a technical limitation of using the Tg(opn1mw2:egfp) line for detection of opn1mw4 expression in the GFP-positive cells.
The manuscript lacks a description of the sampling time point. It is well known that many genes are expressed with daily (or circadian) fluctuation (cf. Doherty & Kay, 2010 Annu. Rev. Genet.). For example, the cone-specific gene list in Fig.2C includes a circadian clock gene, per3, whose expression was reported to fluctuate in a circadian manner in many tissues of zebrafish including the retina (Kaneko et al. 2006 PNAS). It appears to be cone-specific at this time point of sample collection as shown in Fig.2, but might be expressed in a different pattern at other time points (eg, rod expression). The authors should add, at least, a clear description of the sampling time points so as to make their data more informative.
-