Delineating the transcriptional landscape and clonal diversity of virus-specific CD4+ T cells during chronic viral infection
Curation statements for this article:-
Curated by eLife
Evaluation Summary:
This study by Zander et al provides a valuable transcriptomic resource of murine CD4 T cell subsets in chronic viral infection. This study will be of broad interest to a wide range of researchers focused on studying CD4 T cell biology.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. The reviewers remained anonymous to the authors.)
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Although recent evidence indicates that CD4 + T cells responding to chronic viral infection are functionally heterogenous, our understanding of the developmental relationships between these subsets, and a determination of how their transcriptional landscape compares to their acute infection counterparts remains unclear. Additionally, whether cell-intrinsic factors such as TCR usage influence CD4 + T cell fate commitment during persistent infection has not previously been studied. Herein, we perform single-cell RNA sequencing (scRNA-seq) combined with single-cell T cell receptor sequencing (scTCR-seq) on virus-specific CD4 + T cells isolated from mice infected with chronic lymphocytic choriomeningitis virus (LCMV) infection. We identify several transcriptionally distinct states among the Th1, Tfh, and memory-like T cell subsets that form at the peak of infection, including the presence of a previously unrecognized Slamf7 + subset with cytolytic features. We further show that the relative distribution of these populations differs substantially between acute and persistent LCMV infection. Moreover, while the progeny of most T cell clones displays membership within each of these transcriptionally unique populations, overall supporting a one cell-multiple fate model, a small fraction of clones display a biased cell fate decision, suggesting that TCR usage may impact CD4 + T cell development during chronic infection. Importantly, comparative analyses further reveal both subset-specific and core gene expression programs that are differentially regulated between CD4 + T cells responding to acute and chronic LCMV infection. Together, these data may serve as a useful framework and allow for a detailed interrogation into the clonal distribution and transcriptional circuits underlying CD4 + T cell differentiation during chronic viral infection.
Article activity feed
-

-

Author Response
Reviewer #1 (Public Review):
The present study by Zander et al. aims at improving our understanding of CD4+ T cell heterogeneity in response to chronic viral infections. The authors utilize the murine LCMV c13 infection model and perform single cell RNA seq analysis on day 10 post infection to identify multiple, previously unappreciated, T cell subsets. The authors then go on and verify these analyses using multi-color flow cytometry before comparing the transcriptome of CD4 T cells from chronic infection to a previously generated data set of CD4 T cells obtained from acutely-resolved LCMV infection.
The analyses are very well done and provide some interesting novel insights. In particular, the comparison of CD4 T cell subsets across acute and chronic infections is very exciting as they provide a very valuable …
Author Response
Reviewer #1 (Public Review):
The present study by Zander et al. aims at improving our understanding of CD4+ T cell heterogeneity in response to chronic viral infections. The authors utilize the murine LCMV c13 infection model and perform single cell RNA seq analysis on day 10 post infection to identify multiple, previously unappreciated, T cell subsets. The authors then go on and verify these analyses using multi-color flow cytometry before comparing the transcriptome of CD4 T cells from chronic infection to a previously generated data set of CD4 T cells obtained from acutely-resolved LCMV infection.
The analyses are very well done and provide some interesting novel insights. In particular, the comparison of CD4 T cell subsets across acute and chronic infections is very exciting as they provide a very valuable platform that can answer a long-standing question: do CD4 T cells in chronic infection undergo exhaustion similar to CD8 T cells. While this has been proposed for an extended period, this new dataset by Zander et al. can provide some novel insights by comparing individual cell subsets cross-infection. The manuscript would, however, benefit from a more extensive analysis and focus on this interesting point.
We thank the reviewer for their time and careful assessment of our manuscript. We were happy to hear that the reviewer found our work interesting.
On that note, the authors should take advantage of more accurate and present gene datasets to compare the 'dysfunctional' state of CD4 T cells in chronic infection vs acute infection. Also, a different illustration to demonstrate the module score analyses would be more intuitive.
We have now included T cell “exhaustion” genesets from recently published data (Zander et. al 2019 Immunity), and we have also displayed the relative expression of select signature genes from these genesets in an updated supplemental figure 3.
Also, at multiple sections in the manuscript, the authors are missing the accurate citations as they are still mentioned as '(Ref)'.
We apologize for this oversight and have corrected these citations.
Nevertheless, this study does not require major revisions.
Reviewer #2 (Public Review):
In their study "Delineating the transcriptional landscape and clonal diversity of virus-specific CD4+ T cells during chronic viral infection" Zander and co-workers analyze the phenotypic and clonotypic distributions of T cells specific to a LCMV epitope following infection with a chronic LCMV strain in mice. The paper largely follows an earlier study from the same group (Khatun JEM 2021) that has used a similar experimental strategy to analyze T cells responding to an LCMV strain establishing acute infection, and it adds a scTCRseq component to another earlier study of chronic LCMV (Zander Immunity 2022). The main contributions of the paper are to demonstrate that interesting differences between gene expression profiles between chronic and acute LCMV exist, and to identify a new T cell subset (of unknown functional significance).
While the paper is framed around differences between T cell responses to acute and chronic infections, all analysis is done on T cells at day 10 post primary infection. At such an early time point even the acute LCMV strain virus is likely not completely cleared, or at the very least viral antigens are still presented. The relevance of the presented phenotypic differences to other settings with long-term chronic infection is thus questionable. Additionally, there are a number of methodological concerns regarding the robustness of the statistical and bioinformatic analyses that put in doubt some of the conclusions. Most notably, the analysis of fate biases needs to be substantiated by tests against baseline expectations from random assortment to test for statistical significance.
We thank the reviewer for their careful review of our manuscript as well as their helpful comments.
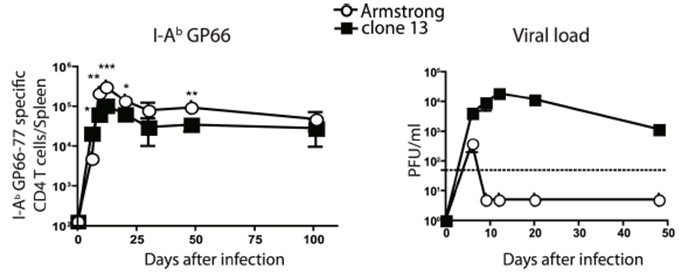
Regarding the day 10 time point-post LCMV Armstrong infection, several groups have previously reported that LCMV viral load is undetectable by day 10 post-infection (see one published example below), although we completely agree with the reviewer that there is still likely to be viral antigens being presented at this time point, as well as ongoing inflammation, which we believe (and as discussed further below) is actually a strength of the study as it allows for a more fair comparison of the transcriptional state of recently stimulated virus-specific CD4 T cells under different contexts (acute vs chronic LCMV infection) . We chose day 10 post LCMV Cl13 and LCMV Armstrong infections as the timepoint for analysis, as this is approximately the peak of the endogenous Gp66-77 CD4+ T cell response (see previously published data below), and is also when there is a more balanced distribution of Th1, Tfh, and T central memory precursor (Tcmp)/ or memory-like cells in these settings, thereby allowing for sufficient numbers of cells/cluster to conduct an in-depth analysis and high-resolution comparison of these subsets between the two different infections. Further, as some degree of TCR stimulation is still likely being experienced at this timepoint during LCMV Armstrong infection, we believe that this is a more useful comparison than at a memory time point (when CD4 T cells are in a quiescent state) as it gives us a better picture of the differentially expressed genes at the peak of the CD4 T cell response, and also provides insight into how chronic viral infection perturbs the transcriptional program of CD4 T cells.
-

Evaluation Summary:
This study by Zander et al provides a valuable transcriptomic resource of murine CD4 T cell subsets in chronic viral infection. This study will be of broad interest to a wide range of researchers focused on studying CD4 T cell biology.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. The reviewers remained anonymous to the authors.)
-

Reviewer #1 (Public Review):
The present study by Zander et al. aims at improving our understanding of CD4+ T cell heterogeneity in response to chronic viral infections. The authors utilize the murine LCMV c13 infection model and perform single cell RNA seq analysis on day 10 post infection to identify multiple, previously unappreciated, T cell subsets. The authors then go on and verify these analyses using multi-color flow cytometry before comparing the transcriptome of CD4 T cells from chronic infection to a previously generated data set of CD4 T cells obtained from acutely-resolved LCMV infection.
The analyses are very well done and provide some interesting novel insights. In particular, the comparison of CD4 T cell subsets across acute and chronic infections is very exciting as they provide a very valuable platform that can answer a …
Reviewer #1 (Public Review):
The present study by Zander et al. aims at improving our understanding of CD4+ T cell heterogeneity in response to chronic viral infections. The authors utilize the murine LCMV c13 infection model and perform single cell RNA seq analysis on day 10 post infection to identify multiple, previously unappreciated, T cell subsets. The authors then go on and verify these analyses using multi-color flow cytometry before comparing the transcriptome of CD4 T cells from chronic infection to a previously generated data set of CD4 T cells obtained from acutely-resolved LCMV infection.
The analyses are very well done and provide some interesting novel insights. In particular, the comparison of CD4 T cell subsets across acute and chronic infections is very exciting as they provide a very valuable platform that can answer a long-standing question: do CD4 T cells in chronic infection undergo exhaustion similar to CD8 T cells. While this has been proposed for an extended period, this new dataset by Zander et al. can provide some novel insights by comparing individual cell subsets cross-infection. The manuscript would, however, benefit from a more extensive analysis and focus on this interesting point.
On that note, the authors should take advantage of more accurate and present gene datasets to compare the 'dysfunctional' state of CD4 T cells in chronic infection vs acute infection. Also, a different illustration to demonstrate the module score analyses would be more intuitive.
Also, at multiple sections in the manuscript, the authors are missing the accurate citations as they are still mentioned as '(Ref)'.
Nevertheless, this study does not require major revisions.
-

Reviewer #2 (Public Review):
In their study "Delineating the transcriptional landscape and clonal diversity of virus-specific CD4+ T cells during chronic viral infection" Zander and co-workers analyze the phenotypic and clonotypic distributions of T cells specific to a LCMV epitope following infection with a chronic LCMV strain in mice. The paper largely follows an earlier study from the same group (Khatun JEM 2021) that has used a similar experimental strategy to analyze T cells responding to an LCMV strain establishing acute infection, and it adds a scTCRseq component to another earlier study of chronic LCMV (Zander Immunity 2022). The main contributions of the paper are to demonstrate that interesting differences between gene expression profiles between chronic and acute LCMV exist, and to identify a new T cell subset (of unknown …
Reviewer #2 (Public Review):
In their study "Delineating the transcriptional landscape and clonal diversity of virus-specific CD4+ T cells during chronic viral infection" Zander and co-workers analyze the phenotypic and clonotypic distributions of T cells specific to a LCMV epitope following infection with a chronic LCMV strain in mice. The paper largely follows an earlier study from the same group (Khatun JEM 2021) that has used a similar experimental strategy to analyze T cells responding to an LCMV strain establishing acute infection, and it adds a scTCRseq component to another earlier study of chronic LCMV (Zander Immunity 2022). The main contributions of the paper are to demonstrate that interesting differences between gene expression profiles between chronic and acute LCMV exist, and to identify a new T cell subset (of unknown functional significance).
While the paper is framed around differences between T cell responses to acute and chronic infections, all analysis is done on T cells at day 10 post primary infection. At such an early time point even the acute LCMV strain virus is likely not completely cleared, or at the very least viral antigens are still presented. The relevance of the presented phenotypic differences to other settings with long-term chronic infection is thus questionable. Additionally, there are a number of methodological concerns regarding the robustness of the statistical and bioinformatic analyses that put in doubt some of the conclusions. Most notably, the analysis of fate biases needs to be substantiated by tests against baseline expectations from random assortment to test for statistical significance.
-