Multistep loading of a DNA sliding clamp onto DNA by replication factor C
Curation statements for this article:-
Curated by eLife
Evaluation Summary:
This study is of relevance to the field of DNA replication, describing how an ATPase known as a 'clamp loader' opens a ring-shaped clamp protein and binds DNA to promote the deposition of the clamp around a nucleic acid duplex to support chromosomal replication. The findings on how different regions of the clamp loader bind to and open a clamp, and how the enzyme engages single-stranded and double-stranded regions of target DNAs provide new insights that further our understanding of the clamp loading reaction. It is intriguing that the clamp loader melts the end of the DNA duplex, an activity that had not been observed before or predicted.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. The reviewers remained anonymous to the authors.)
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
The DNA sliding clamp proliferating cell nuclear antigen (PCNA) is an essential co-factor for many eukaryotic DNA metabolic enzymes. PCNA is loaded around DNA by the ATP-dependent clamp loader replication factor C (RFC), which acts at single-stranded (ss)/double-stranded DNA (dsDNA) junctions harboring a recessed 3’ end (3’ ss/dsDNA junctions) and at DNA nicks. To illuminate the loading mechanism we have investigated the structure of RFC:PCNA bound to ATPγS and 3’ ss/dsDNA junctions or nicked DNA using cryogenic electron microscopy. Unexpectedly, we observe open and closed PCNA conformations in the RFC:PCNA:DNA complex, revealing that PCNA can adopt an open, planar conformation that allows direct insertion of dsDNA, and raising the question of whether PCNA ring closure is mechanistically coupled to ATP hydrolysis. By resolving multiple DNA-bound states of RFC:PCNA we observe that partial melting facilitates lateral insertion into the central channel formed by RFC:PCNA. We also resolve the Rfc1 N-terminal domain and demonstrate that its single BRCT domain participates in coordinating DNA prior to insertion into the central RFC channel, which promotes PCNA loading on the lagging strand of replication forks in vitro. Combined, our data suggest a comprehensive and fundamentally revised model for the RFC-catalyzed loading of PCNA onto DNA.
Article activity feed
-

-

Author Response
Reviewer #2 (Public Review):
Schrecker, Castaneda and colleagues present cryo-EM structures of RFC-PCNA bound to 3'ss/dsDNA junction or nicked DNA stabilized by slowly hydrolyzable ATP analogue, ATPyS. They discover that PCNA can adopt an open form that is planar, different from previous models for the loading a sliding clamp. The authors also report a structure with closed PCNA, supporting the notion that closure of the sliding clamp does not require ATP hydrolysis. The structures explain how DNA can be threaded laterally through a gap in the PCNA trimer, as this process is supported by partial melting of the DNA prior to insertion. The authors also visualise and assign a function to the N-terminal domain in the Rfc1 subunit of the clamp loader, which they find modulates PCNA loading at the replication forks, in turn …
Author Response
Reviewer #2 (Public Review):
Schrecker, Castaneda and colleagues present cryo-EM structures of RFC-PCNA bound to 3'ss/dsDNA junction or nicked DNA stabilized by slowly hydrolyzable ATP analogue, ATPyS. They discover that PCNA can adopt an open form that is planar, different from previous models for the loading a sliding clamp. The authors also report a structure with closed PCNA, supporting the notion that closure of the sliding clamp does not require ATP hydrolysis. The structures explain how DNA can be threaded laterally through a gap in the PCNA trimer, as this process is supported by partial melting of the DNA prior to insertion. The authors also visualise and assign a function to the N-terminal domain in the Rfc1 subunit of the clamp loader, which they find modulates PCNA loading at the replication forks, in turn required for processive synthesis and ligation of Okazaki fragments.
This work is extremely well done, with several structures with resolutions better than 3Å, which a significant achievement given the dynamic nature of the PCNA ring loading process. To investigate the role of the N-terminal domain of Rfc1 in PCNA loading, the authors use in vitro reconstitution of the entire DNA replication reaction, which is a powerful method to identify specific defects in Okazaki fragment synthesis and ligation.
Important issues
- Figure 3B,D,F. I would find them much more informative if the authors showed the overlay between atomic model and cryo-EM density in the main figure. If the figure becomes too busy, the authors could decide to just add additional panels with the overlay as well as the atomic models alone. I do not think that showing segmented density for the DNA alone, as done is Figure 6C is sufficient. Also including the density for e.g. residues Trp638 and Phe582 seems important.
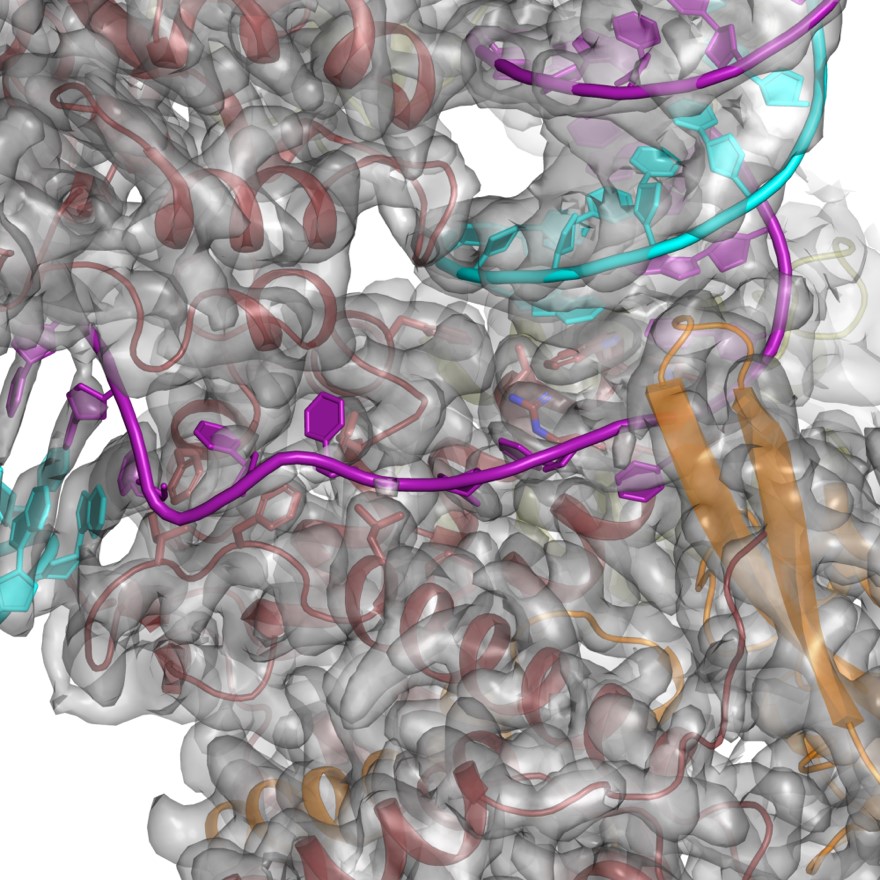
We thank the reviewer for the suggestion. However, we have been unable to establish a way to show the density for both the protein and DNA in a meaningful manner due to the large number of atoms in the fields of view. For an example, please see Figure 1, which corresponds to Figure 3H. To aid the reader, we have revised several of the Figures and Figure Supplements to include density for the DNA.
Consistent with our structures, recent work from the Kelch group has identified Trp638 and Phe582 as facilitating DNA base flipping (Gaubitz et al., 2022a). Despite the role in base flipping, no growth defects were observed in cells in which either of these residues were mutated and thus their functional role and the role of DNA base-flipping remains unclear.
- Cryo-EM samples preparation included substoichiometric RPA, which has been shown to promote DNA loading of PCNA by RFC. Would the authors expect a subset of PCNA-RFC-DNA particles to contain RPA as well? The glycerol gradient gel indicates that, at least in fraction 5, a complex might exist. If the authors think that the particles analyzed cannot contain RPA, it would be useful to mention this.
We have no evidence to suggest that RPA cannot be present in the imaged particles. We have revised the text (lines 150 - 152) clarify that while RPA was present in the sample, we did not observe any density that could not be assigned to either DNA, RFC or PCNA. We therefore suggest that RPA does not interact with the complex in a stable manner.
- Published kinetic data indicate that ATP hydrolysis occurs before clamp closure. To incorporate this notion in their model, the authors suggest that ATP hydrolysis might promote PCNA closure by disrupting the planar RFC:PCNA interaction surface and hence the dynamic interaction of PCNA with Rfc2 and -5 in the open state. In addition, ATP hydrolysis promotes RFC disengagement from PCNA-DNA by reverting from a planar to an out-of-plane state. This model appears reasonable and nicely combines published data with the new findings reported by the authors. However, the model is oversimplified in Figure 6, where the only depicted effect of ATP hydrolysis is RFC release. Perhaps the authors could use the figure caption to acknowledge that ATP hydrolysis likely still has a role in facilitating PCNA closure.
We have revised Figure 6 to show that DNA hydrolysis may occur either before or after ring closure.
- Can the authors explain what steps should be taken to describe PCNA loading by RFC in conditions where ATP hydrolysis is permitted? How would such experiments further inform the molecular mechanism for the loading of the PCNA clamp?
As highlighted in point 3 above and by the other reviewers, ATP and ATPgS may alter the behavior and energetic landscape of RFC. In our studies, ATPgS was added trap the complex in a pre-hydrolysis state in which all components are assembled. We have added a section to the discussion noting the potential differences and highlighting the need for future studies to better elucidate the role of nucleotide hydrolysis. To achieve a hydrolysis competent complex, one could apply time-resolved cryo-EM approaches where the complex is formed on the grids and quickly vitrified. Such an approach, particularly if coupled with stopped-flow kinetic analyses, may provide additional insights in the kinetics of loading of PCNA onto DNA by RFC.
-

Evaluation Summary:
This study is of relevance to the field of DNA replication, describing how an ATPase known as a 'clamp loader' opens a ring-shaped clamp protein and binds DNA to promote the deposition of the clamp around a nucleic acid duplex to support chromosomal replication. The findings on how different regions of the clamp loader bind to and open a clamp, and how the enzyme engages single-stranded and double-stranded regions of target DNAs provide new insights that further our understanding of the clamp loading reaction. It is intriguing that the clamp loader melts the end of the DNA duplex, an activity that had not been observed before or predicted.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the …
Evaluation Summary:
This study is of relevance to the field of DNA replication, describing how an ATPase known as a 'clamp loader' opens a ring-shaped clamp protein and binds DNA to promote the deposition of the clamp around a nucleic acid duplex to support chromosomal replication. The findings on how different regions of the clamp loader bind to and open a clamp, and how the enzyme engages single-stranded and double-stranded regions of target DNAs provide new insights that further our understanding of the clamp loading reaction. It is intriguing that the clamp loader melts the end of the DNA duplex, an activity that had not been observed before or predicted.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. The reviewers remained anonymous to the authors.)
-

Reviewer #1 (Public Review):
In the present manuscript, Schrecker et al. provide a series of structural views of a eukaryotic (budding yeast) clamp loader RFC bound to ATPgS, a client clamp, and substrate DNA. Cryo-electron microscopy is used to image the clamp loader complexes, revealing several different conformational intermediates. In vitro clamp loading, ATPase and replication assays are then employed to test some of the findings resulting from the structural analysis.
The paper is generally well-written and organized in a straightforward manner. The strengths of the work include the high detail (resolution) of the structural models and the multiple conformational intermediates that are observed, which together are used to develop a sensible mechanistic model for clamp loader function that builds on prior work in this system. New …
Reviewer #1 (Public Review):
In the present manuscript, Schrecker et al. provide a series of structural views of a eukaryotic (budding yeast) clamp loader RFC bound to ATPgS, a client clamp, and substrate DNA. Cryo-electron microscopy is used to image the clamp loader complexes, revealing several different conformational intermediates. In vitro clamp loading, ATPase and replication assays are then employed to test some of the findings resulting from the structural analysis.
The paper is generally well-written and organized in a straightforward manner. The strengths of the work include the high detail (resolution) of the structural models and the multiple conformational intermediates that are observed, which together are used to develop a sensible mechanistic model for clamp loader function that builds on prior work in this system. New findings include establishing a role for the BRCT domain of RFC1 in binding DNA, showing how DNA is distorted by clamp loader binding, and showing how the clamp loader can associate with both primer/template and nicked DNA substrates. These insights are well supported by the data shown in the manuscript. The structural data are also used to propose a refinement of a model for clamp loader function in which, contrary to prior published findings, ATP hydrolysis is not required for loaded clamps to close around a duplex DNA. This conclusion is sensible given the structural data, although experimental evidence that the nucleotide substrate used was not hydrolyzed while preparing the sample for imaging by EM is not shown and the article should be revised to provide this evidence.
-

Reviewer #2 (Public Review):
Schrecker, Castaneda and colleagues present cryo-EM structures of RFC-PCNA bound to 3'ss/dsDNA junction or nicked DNA stabilized by slowly hydrolyzable ATP analogue, ATPyS. They discover that PCNA can adopt an open form that is planar, different from previous models for the loading a sliding clamp. The authors also report a structure with closed PCNA, supporting the notion that closure of the sliding clamp does not require ATP hydrolysis. The structures explain how DNA can be threaded laterally through a gap in the PCNA trimer, as this process is supported by partial melting of the DNA prior to insertion. The authors also visualise and assign a function to the N-terminal domain in the Rfc1 subunit of the clamp loader, which they find modulates PCNA loading at the replication forks, in turn required for …
Reviewer #2 (Public Review):
Schrecker, Castaneda and colleagues present cryo-EM structures of RFC-PCNA bound to 3'ss/dsDNA junction or nicked DNA stabilized by slowly hydrolyzable ATP analogue, ATPyS. They discover that PCNA can adopt an open form that is planar, different from previous models for the loading a sliding clamp. The authors also report a structure with closed PCNA, supporting the notion that closure of the sliding clamp does not require ATP hydrolysis. The structures explain how DNA can be threaded laterally through a gap in the PCNA trimer, as this process is supported by partial melting of the DNA prior to insertion. The authors also visualise and assign a function to the N-terminal domain in the Rfc1 subunit of the clamp loader, which they find modulates PCNA loading at the replication forks, in turn required for processive synthesis and ligation of Okazaki fragments.
This work is extremely well done, with several structures with resolutions better than 3Å, which a significant achievement given the dynamic nature of the PCNA ring loading process. To investigate the role of the N-terminal domain of Rfc1 in PCNA loading, the authors use in vitro reconstitution of the entire DNA replication reaction, which is a powerful method to identify specific defects in Okazaki fragment synthesis and ligation.
Important issues
1. Figure 3B,D,F. I would find them much more informative if the authors showed the overlay between atomic model and cryo-EM density in the main figure. If the figure becomes too busy, the authors could decide to just add additional panels with the overlay as well as the atomic models alone. I do not think that showing segmented density for the DNA alone, as done is Figure 6C is sufficient. Also including the density for e.g. residues Trp638 and Phe582 seems important.
2. Cryo-EM samples preparation included substoichiometric RPA, which has been shown to promote DNA loading of PCNA by RFC. Would the authors expect a subset of PCNA-RFC-DNA particles to contain RPA as well? The glycerol gradient gel indicates that, at least in fraction 5, a complex might exist. If the authors think that the particles analyzed cannot contain RPA, it would be useful to mention this.
3. Published kinetic data indicate that ATP hydrolysis occurs before clamp closure. To incorporate this notion in their model, the authors suggest that ATP hydrolysis might promote PCNA closure by disrupting the planar RFC:PCNA interaction surface and hence the dynamic interaction of PCNA with Rfc2 and -5 in the open state. In addition, ATP hydrolysis promotes RFC disengagement from PCNA-DNA by reverting from a planar to an out-of-plane state. This model appears reasonable and nicely combines published data with the new findings reported by the authors. However, the model is oversimplified in Figure 6, where the only depicted effect of ATP hydrolysis is RFC release. Perhaps the authors could use the figure caption to acknowledge that ATP hydrolysis likely still has a role in facilitating PCNA closure.
4. Can the authors explain what steps should be taken to describe PCNA loading by RFC in conditions where ATP hydrolysis is permitted? How would such experiments further inform the molecular mechanism for the loading of the PCNA clamp?
-

Reviewer #3 (Public Review):
In this report, Schrecker et al. use cryo-EM to examine structures of ternary complexes containing the S. cerevisiae clamp loader (RFC), sliding clamp (PCNA), and two different DNA molecules, a DNA molecule with a ss/ds DNA junction with a 3' recessed end (3' ss/ds DNA) and a nicked DNA molecule. These are the first structural data for RFC containing the full-length Rfc1 subunit, and these structures along with biochemical assays demonstrate that Rfc1 interacts with dsDNA to increase the efficiency of clamp loading, particularly on the lagging strand. In order for clamp loaders to accomplish the mechanical task of binding clamps, opening clamps, chaperoning clamps to the appropriate DNA sites, and releasing the clamps onto DNA, multiple clamp loader complexes and conformational states must exist. To date, we …
Reviewer #3 (Public Review):
In this report, Schrecker et al. use cryo-EM to examine structures of ternary complexes containing the S. cerevisiae clamp loader (RFC), sliding clamp (PCNA), and two different DNA molecules, a DNA molecule with a ss/ds DNA junction with a 3' recessed end (3' ss/ds DNA) and a nicked DNA molecule. These are the first structural data for RFC containing the full-length Rfc1 subunit, and these structures along with biochemical assays demonstrate that Rfc1 interacts with dsDNA to increase the efficiency of clamp loading, particularly on the lagging strand. In order for clamp loaders to accomplish the mechanical task of binding clamps, opening clamps, chaperoning clamps to the appropriate DNA sites, and releasing the clamps onto DNA, multiple clamp loader complexes and conformational states must exist. To date, we have a limited structural view of these complexes and conformational states that is based on visualization of a handful of structures from several different organisms. Importantly in this work, the authors were able to capture three different RFC-PCNA-DNA complexes with proteins from the same organism. And this series of structures provides key insight into the structural basis by which RFC-PCNA initially binds DNA and passes ds DNA through the opening of PCNA into the central chamber of the clamp loader. The data show that RFC opens PCNA wide enough to allow dsDNA to pass through the opening which differs from models based on structures from other organisms and molecular dynamics simulations. Unexpectedly, PCNA is open in a planar configuration in a geometry that resembles a horseshoe rather than opening out-of-plane in a spiral configuration. Another intriguing surprise is that RFC melts several base pairs at the primer 3' end, but it is not yet clear how this may contribute to DNA binding or specificity for 3' ss/ds DNA. One potential caveat to these studies is that ATPgammaS was substituted for ATP to block hydrolysis and trap intermediate complexes. It is possible that either RFC conformations or the relative populations of different conformational states are influenced by the bound nucleotide. Overall, this is an important study that answers many questions about the mechanism of clamp loading and also raises some intriguing new questions to stimulate further studies.
-