Vision-related convergent gene losses reveal SERPINE3’s unknown role in the eye
Curation statements for this article:-
Curated by eLife
Evaluation Summary:
The authors use a comparative genomics approach to predict gene function, in particular genes that have a role in eye development. After identifying the convergent loss of SERPINE3 with vision loss across mammals, the authors confirmed its involvement in eye development by characterizing zebrafish knockouts. This work highlights the power of comparative genomics to generate hypotheses that can be experimentally validated. This work is relevant to a broad audience interested in evolution and adaptation as well as for those studying eye development and eye pathologies.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #2 agreed to share their name with the authors.)
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Despite decades of research, knowledge about the genes that are important for development and function of the mammalian eye and are involved in human eye disorders remains incomplete. During mammalian evolution, mammals that naturally exhibit poor vision or regressive eye phenotypes have independently lost many eye-related genes. This provides an opportunity to predict novel eye-related genes based on specific evolutionary gene loss signatures. Building on these observations, we performed a genome-wide screen across 49 mammals for functionally uncharacterized genes that are preferentially lost in species exhibiting lower visual acuity values. The screen uncovered several genes, including SERPINE3 , a putative serine proteinase inhibitor. A detailed investigation of 381 additional mammals revealed that SERPINE3 is independently lost in 18 lineages that typically do not primarily rely on vision, predicting a vision-related function for this gene. To test this, we show that SERPINE3 has the highest expression in eyes of zebrafish and mouse. In the zebrafish retina, serpine3 is expressed in Müller glia cells, a cell type essential for survival and maintenance of the retina. A CRISPR-mediated knockout of serpine3 in zebrafish resulted in alterations in eye shape and defects in retinal layering. Furthermore, two human polymorphisms that are in linkage with SERPINE3 are associated with eye-related traits. Together, these results suggest that SERPINE3 has a role in vertebrate eyes. More generally, by integrating comparative genomics with experiments in model organisms, we show that screens for specific phenotype-associated gene signatures can predict functions of uncharacterized genes.
Article activity feed
-

-

Author Response
Reviewer #2 (Public Review):
We are in a golden age for comparative genomics and this is a prime example of the utility of the field. "Vision-related convergent gene losses reveal SERPINE3's unknown role in the eye" details the discovery of a function for a previously uncharacterized gene in regulating organ development in evolution. The authors intersect patterns of gene loss, quantified as the percentage of intact coding sequence, with visual acuity scores across Mammalia. This analysis identified 26 significant genes that have undergone convergent loss with phenotypic decreases of vision. Many of those genes have previously been annotated in the eye, indicating the analysis was successful and suggesting the uncharacterized genes may also have roles there.
The authors ruled out the top hit due to its specific …
Author Response
Reviewer #2 (Public Review):
We are in a golden age for comparative genomics and this is a prime example of the utility of the field. "Vision-related convergent gene losses reveal SERPINE3's unknown role in the eye" details the discovery of a function for a previously uncharacterized gene in regulating organ development in evolution. The authors intersect patterns of gene loss, quantified as the percentage of intact coding sequence, with visual acuity scores across Mammalia. This analysis identified 26 significant genes that have undergone convergent loss with phenotypic decreases of vision. Many of those genes have previously been annotated in the eye, indicating the analysis was successful and suggesting the uncharacterized genes may also have roles there.
The authors ruled out the top hit due to its specific expression in the testis, and instead performed an in-depth characterization of the second hit, SERPINE3. This included an impressive breadth of comparative genomics across 430 placental mammals, carefully describing the many and diverse genetic perturbations of SERPINE3 in lineages with low visual acuity. These results are persuasive that SERPINE3 is involved in vision, and it is a great example and description of gene loss in adaptation.
Critically, the authors validated the role of SERPINE3 in eye structure by confirming expression patterns in the eye, and characterizing its knockout in zebrafish, demonstrating both qualitative and quantitative impairments to eye structure. This is particularly satisfying as many comparative genomics make such associations but never validate the result. Here, validation of SERPINE3 was an undeniable success and puts a functional annotation to a previously uncharacterized gene. The utility of comparative genomics and zebrafish genetic models has been expertly capitalized upon and there is no doubt our knowledge of eye genetics has increased.
We thank the reviewer for these kind words and the valuable comments that we addressed below.
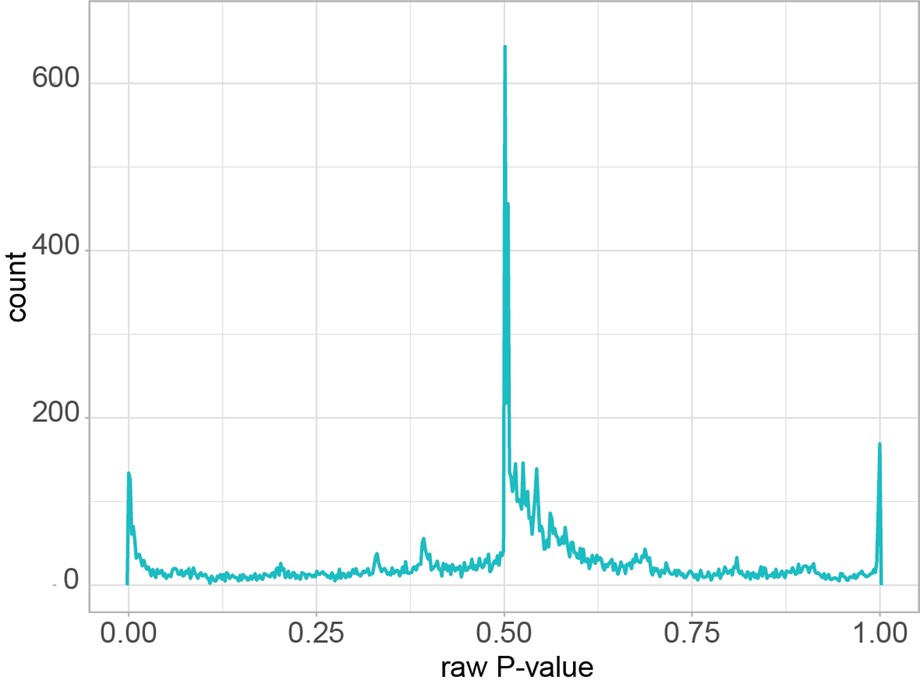
While these end results are certainly valuable to the community, details regarding the statistics and filters underlying the initial convergence analysis are too sparse to interpret. The impressive false discovery rate of the top hits is called into question when the top hit (corrected p-value < 1.1E-15 with visual acuity < 2) is subsequently skipped due to its specific expression in the testes. Given this disconnect, and without knowing the rationale and consequences of the various filters, it is difficult to get a sense of the accuracy and robustness of these p-values. Plots of p-value distributions across the dataset would demonstrate the method is statistically sound and would provide the backdrop to interpret the top hits of interest.
We have now simplified the workflow to detect convergent gene losses in species with lower visual acuity values and explained the rationale of each step (this is detailed in the responses below). We would like to mention that our screen may find genes that are associated with other phenotypes that are shared between species exhibiting lower visual acuity values. For example, several of these species are subterranean mammals, which share other traits and adaptations to their environment. While we do not know to which trait the loss of the testis gene TSACC is associated with, its FDR is only slightly lower than the FDR of the second-ranked SERPINE3 (FDR 1.1E-6 vs. 1.5E-6).
As suggested, we plotted the distribution of the raw P-values of all 13172 genes for which we ran the phylogenetic least square approach. This distribution has a peak at low P-values, indicating that some genes are preferentially lost in the poor-vision mammals. The distribution also showed a peak at ~0.5 and at ~1. We investigated which patterns of the %intact reading frame values appear to contribute to these two peaks.
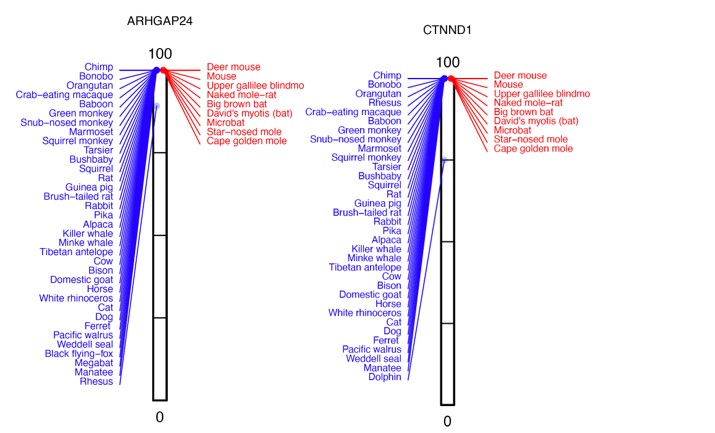
Many genes with P-values of ~0.5 have one high-acuity species (blue), where the %intact value is slightly reduced, whereas other high- and poor-acuity (red) species all have a 100% intact reading frame. Two examples, where rhesus or dolphin have lower %intact values are shown below:
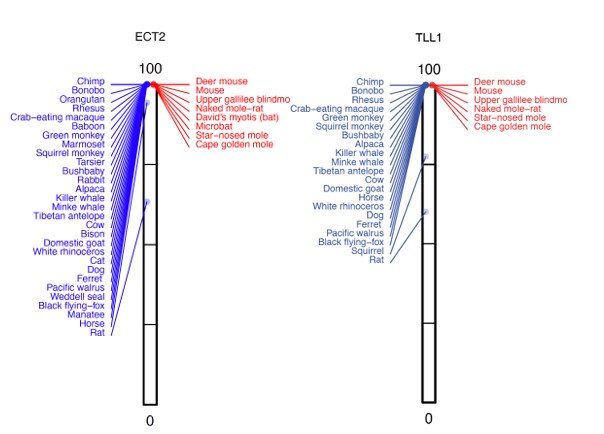
Similarly, many genes with P-values of ~1 have two or more high-acuity species, where the %intact value is reduced, whereas all other species have a 100% intact reading frame.
Since these genes have lower %intact values in a few high-acuity species, the high P-values likely capture a negative association with our trait of interest. While it is not clear why many P-values are around 0.5 or 1, it is clear that these genes are not associated with poor vision.
Our main purpose of using the phylogenetic least square approach was to rank the genes by their association with the poor vision phenotype. Importantly, the top-ranked candidates are all preferentially lost in low-acuity mammals, which is evident from Figure 1A. Furthermore, for SERPINE3, where we experimentally confirmed an eye-related function, three different screens with different phenotype definitions robustly support a preferential loss in low-acuity species (detailed below).
Notes on how many genes pass each filter, and what kinds of genes, would allow interpretation of possible bias in those filters and how they interact with the convergence analysis.
We thank the reviewer for this suggestion. As detailed below, we have now simplified the filtering procedure, justified the filter steps in the revised methods section, and added a flowchart (Figure 1 - supplement figure 1) describing each step and how many genes passed each filter (below).
For instance, the slight changes in visual acuity cutoffs have non-obvious operational consequences for vision, yet large impacts on the resulting gene lists, making it difficult to interpret how the measure is functioning. Most importantly, a negative control in the convergence analysis, demonstrating a null p-value distribution with the same filters, would assuage most concerns.
The reviewer is correct that changes in the visual acuity cutoff leads to different gene lists because the screen searches for genes preferentially lost in different species. However, our screens using three visual acuity cutoffs consistently find SERPINE3 as a candidate in the top 8 genes (Figure 1 - source data 5,6), showing that the association with lower visual acuity is robust for this gene.
As suggested, we have now run a negative control screen. For the negative control, we considered close relatives of the low-acuity species as trait-loss species. Specifically, we selected elephant, rhinoceros, horse, the two flying foxes, guinea pig, degu and squirrel. These 8 species represent five independent lineages. All other species (including the low-acuity species) were treated as trait-preserving species. A Forward genomics screen with otherwise identical filter parameters retrieved only two hits, TUBAL3 and TRIM52, which have no known function in the eye. This supports the specificity of our screen.
We added this to the main text:
“To confirm the specificity of these results, we performed a control screen for genes that are preferentially lost in high-acuity sister species of the low-acuity mammals. This control screen retrieved only two genes, none of which have known functions in the eye (Figure 1 - source data 4). Together, this shows that our genome-wide screen for genes preferentially lost in low-acuity species successfully retrieved known vision-related genes.”
and Methods:
“As a control to ensure that a Forward Genomics screen does not always retrieve vision-related genes, we ran a new screen, searching for genes preferentially lost in high-acuity sister species (elephant, rhinoceros, horse, two flying foxes, guinea pig, degu, squirrel) of the low-acuity mammals that we used in the original screen. All other species including the other high-acuity mammals were then treated as background (Figure 1 - source data 4).“
-

Evaluation Summary:
The authors use a comparative genomics approach to predict gene function, in particular genes that have a role in eye development. After identifying the convergent loss of SERPINE3 with vision loss across mammals, the authors confirmed its involvement in eye development by characterizing zebrafish knockouts. This work highlights the power of comparative genomics to generate hypotheses that can be experimentally validated. This work is relevant to a broad audience interested in evolution and adaptation as well as for those studying eye development and eye pathologies.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #2 agreed to share their name with the authors.)
-

Reviewer #1 (Public Review):
The manuscript by Hiller et al. uses a comparative genomics approach to uncover a previously unknown role of SERPINE3 in eye anatomy. The authors take advantage of available genomes for a variety of mammalian species and search the genomes for evidence of loss of functions or relaxed selection in genes shared in species with low visual acuity (e.g., species that live underground, are nocturnal or mainly rely on echolocation). They identify 26 genes of which many have known functions in eye development or function, but also a handful that have previously not been described to have a role in eye function. The authors then focus on one of these genes (serpine3) as a proof of principle and study expression and function of serpine3 in zebrafish. They find that the gene is expressed in the retina and that knocking …
Reviewer #1 (Public Review):
The manuscript by Hiller et al. uses a comparative genomics approach to uncover a previously unknown role of SERPINE3 in eye anatomy. The authors take advantage of available genomes for a variety of mammalian species and search the genomes for evidence of loss of functions or relaxed selection in genes shared in species with low visual acuity (e.g., species that live underground, are nocturnal or mainly rely on echolocation). They identify 26 genes of which many have known functions in eye development or function, but also a handful that have previously not been described to have a role in eye function. The authors then focus on one of these genes (serpine3) as a proof of principle and study expression and function of serpine3 in zebrafish. They find that the gene is expressed in the retina and that knocking out this gene leads to morphological alterations in the zebrafish eye. Finally, they identified SNPs nearby serpine3 in humans that are linked to eye disorders.
Taken together, the study shows that it is possible to take advantage of evolutionary divergence patterns to reveal novel insights into gene function.
What is exciting about this paper is that it uses evolutionary knowledge and comparative approaches to uncover a novel role for a gene that would not necessarily have been discovered without it. While not completely unique, this is rather rare in evolutionary studies which usually rely on knowledge from other systems to uncover candidate genes involved in evolution.
-

Reviewer #2 (Public Review):
We are in a golden age for comparative genomics and this is a prime example of the utility of the field. "Vision-related convergent gene losses reveal SERPINE3's unknown role in the eye" details the discovery of a function for a previously uncharacterized gene in regulating organ development in evolution. The authors intersect patterns of gene loss, quantified as the percentage of intact coding sequence, with visual acuity scores across Mammalia. This analysis identified 26 significant genes that have undergone convergent loss with phenotypic decreases of vision. Many of those genes have previously been annotated in the eye, indicating the analysis was successful and suggesting the uncharacterized genes may also have roles there.
The authors ruled out the top hit due to its specific expression in the testis, …
Reviewer #2 (Public Review):
We are in a golden age for comparative genomics and this is a prime example of the utility of the field. "Vision-related convergent gene losses reveal SERPINE3's unknown role in the eye" details the discovery of a function for a previously uncharacterized gene in regulating organ development in evolution. The authors intersect patterns of gene loss, quantified as the percentage of intact coding sequence, with visual acuity scores across Mammalia. This analysis identified 26 significant genes that have undergone convergent loss with phenotypic decreases of vision. Many of those genes have previously been annotated in the eye, indicating the analysis was successful and suggesting the uncharacterized genes may also have roles there.
The authors ruled out the top hit due to its specific expression in the testis, and instead performed an in-depth characterization of the second hit, SERPINE3. This included an impressive breadth of comparative genomics across 430 placental mammals, carefully describing the many and diverse genetic perturbations of SERPINE3 in lineages with low visual acuity. These results are persuasive that SERPINE3 is involved in vision, and it is a great example and description of gene loss in adaptation.
Critically, the authors validated the role of SERPINE3 in eye structure by confirming expression patterns in the eye, and characterizing its knockout in zebrafish, demonstrating both qualitative and quantitative impairments to eye structure. This is particularly satisfying as many comparative genomics make such associations but never validate the result. Here, validation of SERPINE3 was an undeniable success and puts a functional annotation to a previously uncharacterized gene. The utility of comparative genomics and zebrafish genetic models has been expertly capitalized upon and there is no doubt our knowledge of eye genetics has increased.
While these end results are certainly valuable to the community, details regarding the statistics and filters underlying the initial convergence analysis are too sparse to interpret. The impressive false discovery rate of the top hits is called into question when the top hit (corrected p-value < 1.1E-15 with visual acuity < 2) is subsequently skipped due to its specific expression in the testes. Given this disconnect, and without knowing the rationale and consequences of the various filters, it is difficult to get a sense of the accuracy and robustness of these p-values. Plots of p-value distributions across the dataset would demonstrate the method is statistically sound and would provide the backdrop to interpret the top hits of interest. Notes on how many genes pass each filter, and what kinds of genes, would allow interpretation of possible bias in those filters and how they interact with the convergence analysis. For instance, the slight changes in visual acuity cutoffs have non-obvious operational consequences for vision, yet large impacts on the resulting gene lists, making it difficult to interpret how the measure is functioning. Most importantly, a negative control in the convergence analysis, demonstrating a null p-value distribution with the same filters, would assuage most concerns.
With that said, the primary conclusion that SERPINE3 is involved in eye structure is strong. The extended mining of orthologues across new genomes is impressive, the zebrafish characterization is compelling, and readers should have no doubt SERPINE3 is involved in eye development. These results will be valuable for research into eye disease, understanding eye development, and unraveling the evolution of vision loss in diverse models. This paper will be a reference for all future investigations in these areas and oft cited.
-