Closely related type II-C Cas9 orthologs recognize diverse PAMs
Curation statements for this article:-
Curated by eLife
Evaluation Summary:
This work is relevant to all who are interested in genome editing. The versatile Cas9 nuclease has enabled creative genome editing applications, yet the targetable sequence space is limited by the PAM specificity of the Cas9 RNP. This manuscript expands the Cas9 toolbox by defining the PAM specificity and genome editing activity of a large group of smaller-sized type II-C Cas9s. The results also contribute to our understanding of the diversity of Cas enzymes and show that there is a significant potential in mining for non-trivial genome editing tools amongst highly similar Cas orthologs.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #3 agreed to share their name with the authors.)
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
The RNA-guided CRISPR/Cas9 system is a powerful tool for genome editing, but its targeting scope is limited by the protospacer-adjacent motif (PAM). To expand the target scope, it is crucial to develop a CRISPR toolbox capable of recognizing multiple PAMs. Here, using a GFP-activation assay, we tested the activities of 29 type II-C orthologs closely related to Nme1Cas9, 25 of which are active in human cells. These orthologs recognize diverse PAMs with variable length and nucleotide preference, including purine-rich, pyrimidine-rich, and mixed purine and pyrimidine PAMs. We characterized in depth the activity and specificity of Nsp2Cas9. We also generated a chimeric Cas9 nuclease that recognizes a simple N 4 C PAM, representing the most relaxed PAM preference for compact Cas9s to date. These Cas9 nucleases significantly enhance our ability to perform allele-specific genome editing.
Article activity feed
-

-

Author Response
Reviewer #1 (Public Review):
The tools and approaches in this manuscript are of broad interest, not only to protein engineers but also to the many researchers using genome-editing reagents. However, putting the work in the context of previous research, both through changing the writing and additional experiments, will be critical for taking advantage of that widespread applicability.
Strengths:
Overall, the data support the conclusions of the manuscript.
The most exciting product of this work is an engineered nuclease, Nsp2-SmuCas9, that has high activity and specificity in human cells and a relaxed PAM preference for a single C base. This chimeric enzyme can efficiently induce indels at endogenous sites. While other works have presented nucleases with minimal PAM preferences, Nsp2-SmuCas9 is a useful …
Author Response
Reviewer #1 (Public Review):
The tools and approaches in this manuscript are of broad interest, not only to protein engineers but also to the many researchers using genome-editing reagents. However, putting the work in the context of previous research, both through changing the writing and additional experiments, will be critical for taking advantage of that widespread applicability.
Strengths:
Overall, the data support the conclusions of the manuscript.
The most exciting product of this work is an engineered nuclease, Nsp2-SmuCas9, that has high activity and specificity in human cells and a relaxed PAM preference for a single C base. This chimeric enzyme can efficiently induce indels at endogenous sites. While other works have presented nucleases with minimal PAM preferences, Nsp2-SmuCas9 is a useful alternative and may be preferred. It is also more compact than the standard SpCas9, making it appealing for gene therapy applications.
Technologically, the presented approach of screening orthologs for new specificities and making chimeras to achieve further diversity is a good way to develop new genome-editing reagents. The authors used appropriate methods, such as GUIDE-seq, to complete their goals. Extending beyond the GFP-activation assay to determine activity at endogenous targets enhanced the value of the results.
Conceptually, it was important information to the field that proteins with very high sequence identity (93%) can have divergent PAM preferences. Through their engineering, the authors clearly demonstrate the advantage of characterizing such close orthologs with diverse amino acids in the area of PAM recognition.
Weaknesses:
- An overall weakness with the work is that it is not clear how the activity level of the relaxed PAM enzyme, Nsp2-SmuCas9, compares to existing enzymes. Is it much better than the SpCas9 that has almost no PAM preference (SpRY) or the NGN PAM (SpG)? How does it compare to the most commonly used SpCas9 nuclease, which is known to be active in a wide variety of biological contexts? The activity assessment at endogenous sites seemed to have a long timeline, as the indel rate was measured 5 days after transfection. Clarifying the effectiveness of this new nuclease would increase the impact of this work.
We sincerely thank the reviewer for the constructive comments on our manuscript. Following reviewer’s suggestions, we compared the editing efficiency of Nsp2Cas9, Nsp2-SmuCas9, SpCas9, SpCas9-NG, and SpCas9-RY side-by-side. Overall, the editing efficiency was low this time probably due to low transfection efficiency. The results revealed that SpCas9 was the most active enzyme. Nsp2Cas9, SpCas9-NG, and SpCas9-RY displayed similar activity. Nsp2-SmuCas9 displayed lower activities than other Cas9 variants (Figure 5C).
- In the presentation of the manuscript, there are several weaknesses. First, while it is true that allele-specific disruption is an important application of new CRISPR proteins, there are many other reasons why they would be useful. The specific focus on this single application throughout the abstract, introduction and discussion takes away from the widespread utility of these new tools. The writing would be more compelling if it targeted a broader audience. Allele-specific targeting is also possible beyond the PAM site if the mutation is in a position with high specificity.
Many thanks for the reviewer’s suggestions. Following reviewer’s suggestions, we emphasize the widespread utility of these new tools throughout the abstract, introduction, and discussion in the revised manuscript. Allele-specific targeting is only mentioned in the discussion.
- Second, the introduction is further missing a discussion of other research engineering new PAM specificity or even completely removing specificity. A more convincing narrative would include reasoning for why characterizing naturally occurring orthologs is a powerful and important approach. This information is in the discussion, but it would be helpful for the reader if these points were in the introduction.
Many thanks for the reviewer’s comments. Following reviewer’s suggestions, we added other research engineering new PAM specificity in the introduction. We also included reasoning for why characterizing naturally occurring orthologs is a powerful and important approach.
“Engineered Cas9 variants with flexible PAMs can increase targeting scope. For example, SaCas9 was engineered to accept an NNNRRT PAM [1]; SpCas9 was engineered to accept almost all PAMs [2], but this strategy is time-consuming, and often comes at a cost of reduced on-target activity. Another strategy is to harness natural Cas9 nucleases for genome editing. We have developed several closely related Cas9 orthologs for genome editing [3, 4]. The advantage of developing tools from closely related Cas9 orthologs is that they can exchange the PAM-interacting (PI) domain. If an ortholog recognizes a particular PAM but does not work efficiently in human cells, we can use this ortholog PI to replace another ortholog PI to generate a chimeric Cas9.”
- A second concern with the presentation and analysis of the findings is a minimal connection to the structural context of the discoveries. Many readers will likely be interested in how the specificity shifts are occurring in these orthologs, which could be remedied by supplementary figures of homology models.
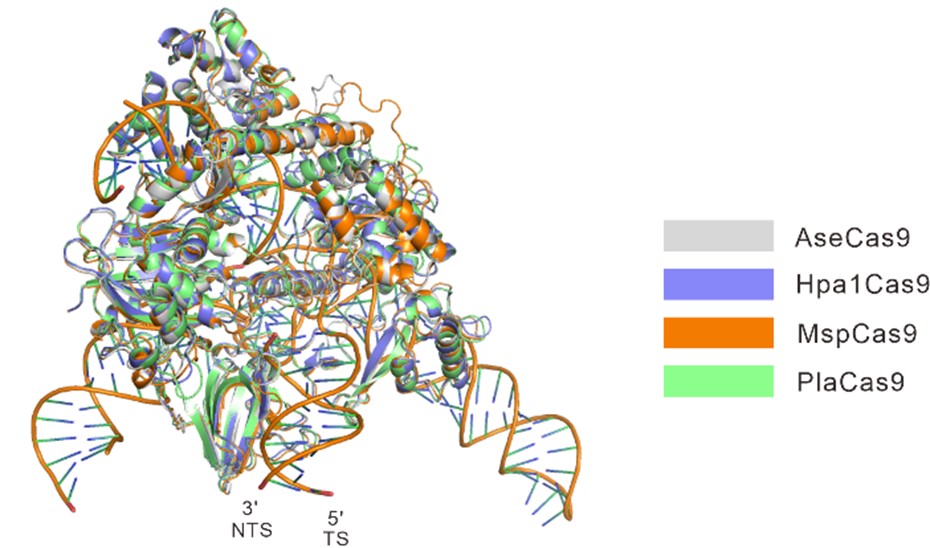
We totally agree with the referee that structural models would help readers better understand the specificity shifts occurring in these orthologs. We have generated calculated structural models of these orthologs in complex with sgRNA and DNA using the crystal structure of Nme1Cas9 (PDB ID: 6JDV). Some specificity shifts can be well explained by these structural models. When the amino acid near the 5 position of the PAM is histidine, its side chain forms a potential hydrogen bond with the 6-hydroxyl group of guanine. Replacement of this guanine by cytosine or thymine would cause a major clash, whereas adenine lacks the hydroxyl group to form hydrogen bond with the histidine (Figure 2-figure supplement 2A). Likewise, an aspartate at 5 position of the PAM would favor a specific recognition of cytosine via hydrogen bonding with its 4-amine group, but not of other bases that may either result in major clash or abolish the hydrogen bond (Figure 2-figure supplement 2B). Similar explanation applies also to the apparent specificity between glutamine and adenine at the 8 position of the PAM on the target sequence (Figure 2-figure supplement 2C).
- Along the same lines, further structural analysis of the failures would be helpful for those embarking on similar projects. Are there any differences in the sequence or structure of the 4/29 orthologs that were not functional in the GFP-activation assay compared to those that were?
Sequence alignment indicates that the four inactive orthologs possess intact active sites. In the predicted structural models of these orthologs, we did not observe local conformational variations that preclude the interaction with sgRNA or DNA. Sequence alignment indicates that the four inactive orthologs possess intact active sites. In the predicted structural models of these orthologs, we did not observe local conformational variations that preclude the interaction with sgRNA or DNA. We speculate that specific modifications of Cas9s in mammalian cells may occur, leading to the loss of enzymatic activities of the 4 orthologs.
Calculated structural models of AseCas9, Hpa1Cas9, MspCas9, and PlaCas9. Overall calculated structures of AseCas9, Hpa1Cas9, MspCas9, and PlaCas9 with sgRNA and dsDNA.
- Similarly, it was surprising that the Nsp2-NarCas9 chimera was not active, and it would be helpful if the authors could speculate based on the differences between SmuCas9 and NarCas9, such as at the interface of the domains that were fused. Structural models of the fusions would help the reader to visualize the strategy. Exploring the failures and challenges is important for understanding the generalizability of the presented approach.
Following reviewer’s comments, we generated structural models of Nsp2-NarCas9, Nsp2-SmuCas9, and NarCas9 using the crystal structure of highly homologous Nme1Cas9 in complex with sgRNA and dsDNA (PDB ID: 6JDV) as the template by SWISS-MODEL. By superimposing these models, we noticed that residues G1035, K1037 and T1038 of Nsp2-NarCas9 chimera protrude towards the DNA molecule, which would prevent the binding with DNA and thereby abolishing the editing activity (Figure 4-figure supplement 2A). In comparison, Nsp2-SmuCas9 and NarCas9, which possess the Cas activity, show no protrusion at the corresponding position (Figure 4-figure supplement 2B-C).
- Finally, the final sequence of Nsp2-SmuCas9 fusion, as well as other enzymes such as the failed Nsp2-NarCas9, are not obvious in the manuscript. I may have missed them, but I also did not see the primers used in the Methods section. Addgene submission is also encouraged and would be of great value to the scientific community.
Thank you for your suggestions. The final sequence of Nsp2-SmuCas9, as well as other enzymes, have been provided in Supplemental file 1. The primers for chimera proteins were listed in Supplemental file 1. We will submit plasmids to Addgene soon.
-

Evaluation Summary:
This work is relevant to all who are interested in genome editing. The versatile Cas9 nuclease has enabled creative genome editing applications, yet the targetable sequence space is limited by the PAM specificity of the Cas9 RNP. This manuscript expands the Cas9 toolbox by defining the PAM specificity and genome editing activity of a large group of smaller-sized type II-C Cas9s. The results also contribute to our understanding of the diversity of Cas enzymes and show that there is a significant potential in mining for non-trivial genome editing tools amongst highly similar Cas orthologs.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #3 agreed to share their name with the …
Evaluation Summary:
This work is relevant to all who are interested in genome editing. The versatile Cas9 nuclease has enabled creative genome editing applications, yet the targetable sequence space is limited by the PAM specificity of the Cas9 RNP. This manuscript expands the Cas9 toolbox by defining the PAM specificity and genome editing activity of a large group of smaller-sized type II-C Cas9s. The results also contribute to our understanding of the diversity of Cas enzymes and show that there is a significant potential in mining for non-trivial genome editing tools amongst highly similar Cas orthologs.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #3 agreed to share their name with the authors.)
-

Reviewer #1 (Public Review):
The tools and approaches in this manuscript are of broad interest, not only to protein engineers but also to the many researchers using genome-editing reagents. However, putting the work in the context of previous research, both through changing the writing and additional experiments, will be critical for taking advantage of that widespread applicability.
Strengths:
Overall, the data support the conclusions of the manuscript.
The most exciting product of this work is an engineered nuclease, Nsp2-SmuCas9, that has high activity and specificity in human cells and a relaxed PAM preference for a single C base. This chimeric enzyme can efficiently induce indels at endogenous sites. While other works have presented nucleases with minimal PAM preferences, Nsp2-SmuCas9 is a useful alternative and may be preferred. …
Reviewer #1 (Public Review):
The tools and approaches in this manuscript are of broad interest, not only to protein engineers but also to the many researchers using genome-editing reagents. However, putting the work in the context of previous research, both through changing the writing and additional experiments, will be critical for taking advantage of that widespread applicability.
Strengths:
Overall, the data support the conclusions of the manuscript.
The most exciting product of this work is an engineered nuclease, Nsp2-SmuCas9, that has high activity and specificity in human cells and a relaxed PAM preference for a single C base. This chimeric enzyme can efficiently induce indels at endogenous sites. While other works have presented nucleases with minimal PAM preferences, Nsp2-SmuCas9 is a useful alternative and may be preferred. It is also more compact than the standard SpCas9, making it appealing for gene therapy applications.
Technologically, the presented approach of screening orthologs for new specificities and making chimeras to achieve further diversity is a good way to develop new genome-editing reagents. The authors used appropriate methods, such as GUIDE-seq, to complete their goals. Extending beyond the GFP-activation assay to determine activity at endogenous targets enhanced the value of the results.
Conceptually, it was important information to the field that proteins with very high sequence identity (93%) can have divergent PAM preferences. Through their engineering, the authors clearly demonstrate the advantage of characterizing such close orthologs with diverse amino acids in the area of PAM recognition.
Weaknesses:
An overall weakness with the work is that it is not clear how the activity level of the relaxed PAM enzyme, Nsp2-SmuCas9, compares to existing enzymes. Is it much better than the SpCas9 that has almost no PAM preference (SpRY) or the NGN PAM (SpG)? How does it compare to the most commonly used SpCas9 nuclease, which is known to be active in a wide variety of biological contexts? The activity assessment at endogenous sites seemed to have a long timeline, as the indel rate was measured 5 days after transfection. Clarifying the effectiveness of this new nuclease would increase the impact of this work.
In the presentation of the manuscript, there are several weaknesses. First, while it is true that allele-specific disruption is an important application of new CRISPR proteins, there are many other reasons why they would be useful. The specific focus on this single application throughout the abstract, introduction and discussion takes away from the widespread utility of these new tools. The writing would be more compelling if it targeted a broader audience. Allele-specific targeting is also possible beyond the PAM site if the mutation is in a position with high specificity. Second, the introduction is further missing a discussion of other research engineering new PAM specificity or even completely removing specificity. A more convincing narrative would include reasoning for why characterizing naturally occurring orthologs is a powerful and important approach. This information is in the discussion, but it would be helpful for the reader if these points were in the introduction.
A second concern with the presentation and analysis of the findings is a minimal connection to the structural context of the discoveries. Many readers will likely be interested in how the specificity shifts are occurring in these orthologs, which could be remedied by supplementary figures of homology models. Along the same lines, further structural analysis of the failures would be helpful for those embarking on similar projects. Are there any differences in the sequence or structure of the 4/29 orthologs that were not functional in the GFP-activation assay compared to those that were? Similarly, it was surprising that the Nsp2-NarCas9 chimera was not active, and it would be helpful if the authors could speculate based on the differences between SmuCas9 and NarCas9, such as at the interface of the domains that were fused. Structural models of the fusions would help the reader to visualize the strategy. Exploring the failures and challenges is important for understanding the generalizability of the presented approach.
Finally, the final sequence of Nsp2-SmuCas9 fusion, as well as other enzymes such as the failed Nsp2-NarCas9, are not obvious in the manuscript. I may have missed them, but I also did not see the primers used in the Methods section. Addgene submission is also encouraged and would be of great value to the scientific community.
-

Reviewer #2 (Public Review):
The versatile Cas9 has enabled creative genome editing applications. The targetable sequence space is limited by the PAM specificity of the Cas9 RNP. In this manuscript, the authors made a comprehensive attempt to explore the genome editing potential of the under-explored type II-C Cas9s. A positive selection assay was set up to define the PAM specificity of twenty nine different II-C Cas9 homologs. Twenty five of them were active in genome editing in human cells. Their distinct PAM specificities can be rationalized to some extent based on the identity of the key residues in the PAM-interaction domain. The authors then focused on Nsp2Cas9 and a few others. Nsp2Cas9 can be engineered to recognize a single C PAM by replacing the PI domain with that of the SmuCas9. Impressively, the resulting chimera retained …
Reviewer #2 (Public Review):
The versatile Cas9 has enabled creative genome editing applications. The targetable sequence space is limited by the PAM specificity of the Cas9 RNP. In this manuscript, the authors made a comprehensive attempt to explore the genome editing potential of the under-explored type II-C Cas9s. A positive selection assay was set up to define the PAM specificity of twenty nine different II-C Cas9 homologs. Twenty five of them were active in genome editing in human cells. Their distinct PAM specificities can be rationalized to some extent based on the identity of the key residues in the PAM-interaction domain. The authors then focused on Nsp2Cas9 and a few others. Nsp2Cas9 can be engineered to recognize a single C PAM by replacing the PI domain with that of the SmuCas9. Impressively, the resulting chimera retained the same level of editing efficiency. A slight drawback is that the Nsp2Cas9 and its chimera derivative had highly off-targeting activity than the benchmark, the previously characterized Nme2Cas9. Overall, the experiments were expertly performed. The work expands the Cas9 toolbox by defining the PAM specificity and genome editing activity of a large group of smaller-sized type II-C Cas9s. The success in making Cas9-PI chimeras points to one effective approach to expand the targeting space of the existing Cas9s.
-

Reviewer #3 (Public Review):
Searches for new types of CRISPR-Cas nucleases bring back enzymes with unique properties. Characterization of closely related Cas homologues can also be useful and result in discoveries of enzymes with properties that complement those of existing editors. The large-scale characterization of Cas9 orthologs by the Siksnys group, research by the Sontheimer, Zhang, and Doudna groups, as well as work from other laboratories, resulted in characterization of Cas9 enzymes which recognize distinct PAMs and can be used to edit a wide range of genomic targets. The authors of this paper contributed to the effort of expanding the Cas9 toolbox by characterizing BlatCas9 and SauriCas9, two enzymes that may be promising for genome engineering.
The main difficulty with most Cas9 orthologs is their low activity in human …
Reviewer #3 (Public Review):
Searches for new types of CRISPR-Cas nucleases bring back enzymes with unique properties. Characterization of closely related Cas homologues can also be useful and result in discoveries of enzymes with properties that complement those of existing editors. The large-scale characterization of Cas9 orthologs by the Siksnys group, research by the Sontheimer, Zhang, and Doudna groups, as well as work from other laboratories, resulted in characterization of Cas9 enzymes which recognize distinct PAMs and can be used to edit a wide range of genomic targets. The authors of this paper contributed to the effort of expanding the Cas9 toolbox by characterizing BlatCas9 and SauriCas9, two enzymes that may be promising for genome engineering.
The main difficulty with most Cas9 orthologs is their low activity in human cells. In the current work, the authors decided to focus on Cas9 proteins highly similar to NmeCas9, which is highly active in eukaryotes, and search for enzymes with novel PAM requirements. The authors determined PAM sequences for 29 NmeCas9 orthologs through screening experiments in human cells. Direct determination of PAM in eukaryotes makes the results highly valuable in terms of the future use of these nucleases in genome editing, as PAM sequences determined in vitro, in bacteria, and in eukaryotic cells do not fully match sometimes.
The results of this work, along with the data of Gasiunas et al., can be used for developing of structure-based Cas9 PAM prediction tools given the new opportunities of protein structure modelling with AlphaFold2.
It is important to note that the Nme1Cas9 orthologs whose activity was considered by the authors as insufficient based on their PAM screening experiment in human cells, may actually be active and efficient enzymes, but poorly produced.
-