Unconventional conservation reveals structure-function relationships in the synaptonemal complex
Curation statements for this article:-
Curated by eLife
Evaluation Summary:
Although the synaptonemal complex (SC) is an essential, deeply conserved structure that holds meiotic chromosomes together, the constituent proteins evolve exceptionally rapidly. This rapid evolution in turn has hindered the identification of SC proteins based solely on sequence homology. This manuscript overcomes this challenge by developing and validating a clever protein structure-based approach that leverages sequence divergence - rather than sequence conservation - to identify novel SC components.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. The reviewers remained anonymous to the authors.)
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Functional requirements constrain protein evolution, commonly manifesting in a conserved amino acid sequence. Here, we extend this idea to secondary structural features by tracking their conservation in essential meiotic proteins with highly diverged sequences. The synaptonemal complex (SC) is a ~100-nm-wide ladder-like meiotic structure present in all eukaryotic clades, where it aligns parental chromosomes and regulates exchanges between them. Despite the conserved ultrastructure and functions of the SC, SC proteins are highly divergent within Caenorhabditis . However, SC proteins have highly conserved length and coiled-coil domain structure. We found the same unconventional conservation signature in Drosophila and mammals, and used it to identify a novel SC protein in Pristionchus pacificus , Ppa-SYP-1. Our work suggests that coiled-coils play wide-ranging roles in the structure and function of the SC, and more broadly, that expanding sequence analysis beyond measures of per-site similarity can enhance our understanding of protein evolution and function.
Article activity feed
-

-

Author Response:
Reviewer #1 (Public Review):
Kursel et al. examined the evolution of synaptonemal complex proteins in C.elegans. While the sequence of the SC proteins evolved rapidly analysis of the structure of SC central region proteins from Caenorhabditis, Drosophila and mammalian species revealed that the length and placement of the coiled-coil domains, as well as overall protein length, were highly conserved across species. This conservation in the structure of coiled-coil proteins within the SC led to the proposal that the conserved structural parameters of the SC proteins and their coiled-coil domains could be used to identify central region components of the SC in species where components could not be identified on sequence conservation alone. Kursel et al demonstrated their parameters could be used to identify a transverse …
Author Response:
Reviewer #1 (Public Review):
Kursel et al. examined the evolution of synaptonemal complex proteins in C.elegans. While the sequence of the SC proteins evolved rapidly analysis of the structure of SC central region proteins from Caenorhabditis, Drosophila and mammalian species revealed that the length and placement of the coiled-coil domains, as well as overall protein length, were highly conserved across species. This conservation in the structure of coiled-coil proteins within the SC led to the proposal that the conserved structural parameters of the SC proteins and their coiled-coil domains could be used to identify central region components of the SC in species where components could not be identified on sequence conservation alone. Kursel et al demonstrated their parameters could be used to identify a transverse filament protein of the SC in the organism Pristionchus pacificus.
Due to high sequence divergence identifying SC proteins in new model systems has been challenging. The identification by Kursel et al. of potential search parameters to identify these diverged proteins will be useful to the those who work on the synaptonemal complex. This approach has the potential to applicable to other types of proteins that show rapid sequence divergence. As the mammalian, fly, and worm SC proteins all displayed different lengths and placements of their coiled-coil domains within their SC proteins this approach is limited by the availability of related identified sequences to the model organism of interest. Additionally, this approach may still yield multiple candidates that fit the structural parameters which will require additional means to ultimately identify the protein of interest. The data in the manuscript supports the authors' claims of structural conservation within SC proteins but only additional applications of their search methods will reveal how useful it is to search for other types of proteins based on structural features.
We thank the reviewer for their summary and feedback. We hope that with the ever-lowered costs of genome assembly and the expansion of CRISPR/Cas9 gene-editing capabilities, the pipeline we developed will be applicable to more clades and species. We agree that it will be interesting to expand our method beyond the SC. Going forward, we are excited to test whether it will enable us to identify other types of proteins, especially those that are part of condensates. In this light, our finding that centrosomal proteins are also enriched in the same evolutionary class as SC proteins is especially intriguing.
Reviewer #2 (Public Review):
In this article, Kursel and colleagues sought to identify evolutionary features of components of the SC the are evident in the absence of strict amino-acid conservation. After identifying three joint evolutionary properties of SC proteins - conservation of coiled-coil architecture, conservation of length and significant amino acid divergence - they show that these properties can be used to identify unknown SC proteins in divergent species. Overall, their general conclusion is very well supported and they do an excellent job functionally testing their approach by showing that one identified candidate for a novel SC protein in Pristionchus is in fact a component of the SC. In addition to providing new insight into the evolutionary forces that shape the evolution of SC proteins, this article provides new insight into how one might generally identify functionally similar or homologous proteins despite very deep divergence. Thus, this work has broader relevance to molecular evolution and evolution of protein structure.
There are some places where smaller conclusions need more support. In particular, it is not entirely clear that this triple pattern - conservation of coiled-coil architecture, conservation of length and significant amino acid divergence - is broadly applicable to SC components beyond Dipterans and Nematodes. In particular, the pattern is weaker in Eutherian mammals. Some further investigation is needed to claim that the pattern is similar in mammals. In addition, it is not clear if coiled-coil conservation rather than simply having a coiled-coil domain is important as a mark of SC proteins. A comparison of coiled-coil conservation among proteins that have coiled-coil domains would be needed for this conclusion. Finally, there should be some additional clarification that not all nematode SC proteins have a pattern of insertion and deletion that is limited to regions outside of the coil-coil domains.
We thank the reviewer for their appreciation of the broader impacts of our work to molecular evolution and for their suggestions for providing more support for our conclusions. We have addressed each of these points below (1. the evolutionary pattern in mammals, 2. the value of the coiled-coil conservation score, and 3. clarification of the indel analysis).
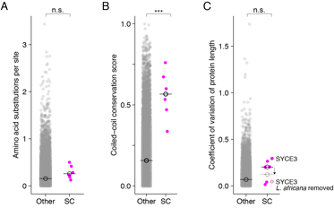
- As suggested, we have added dot plots comparing mammalian SC proteins to all other mammalian proteins for the three metrics central to this manuscript - amino acid substitutions per site, coiled-coil conservation scores and coefficient of variation of protein length. The plots (shown here) can be found in Figure 3 – figure supplement 4.
These plots provide additional evidence that the evolutionary pattern of mammalian SC proteins is similar to (although weaker than) that of Caenorhabitis and Drosophila.
In panel (A), we show the median amino acid substitutions per site of SC proteins is higher than other proteins in mammals, although the difference is not significant. We discuss two reasons why the divergence trend is weaker for mammalian SC proteins in the results. Briefly summarized they are, 1. The overall divergence of the mammalian proteome is less than that of the Caenorhabditis or Drosophila proteome, and 2. Mammalian SC proteins may face additional evolutionary constraints due to novel functions including mammalian-specific protein interactions.
In panel (B), we show that mammalian SC proteins have a significantly higher coiled-coil conservation score than other proteins.
In panel (C), we show coefficient of variation of protein length for mammalian SC proteins is not significantly different than other proteins. We hypothesize that this could be due to gene annotation errors which plague even very high-quality genomes. For example, we found annotation errors in 23 (18%) of the 125 Caenorhabditis SC proteins examined in this study. Uncorrected, these errors often read as large insertions or deletions, and artificially large coefficient of variation. We use L. africana SYCE3 to demonstrate how potential annotation errors could impact our measure of length variation in mammalian SC proteins. L. africana SYCE3 has conspicuous N- and C-terminal extensions not found in any other SYCE3. Excluding that single protein - L. africana SYCE3 – reduces the average length variation from 29% to 4% in the SYCE3 orthogroup, below the median of other proteins. Correspondingly, the median SC coefficient of variation of protein length drops from 20% (unfilled black circle) to 12% (dashed, unfilled circle). While systematic manual annotation of the Eutherian mammals proteomes is beyond the scope of this manuscript, we added in the Discussion explicit reference to the implications of annotation errors on our ability to systematically address evolutionary pressures affecting indels.
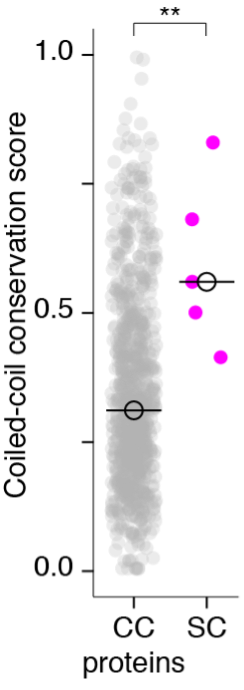
- We thank the reviewers for this important suggestion. Indeed, the inclusion of the few examples in Figure 2 were meant as demonstration rather than a statistical analysis. To create a group of proteins that would serve as appropriate control for conservation of the length and organization of the of coiled-coils, we selected orthogroups in which 90% of the proteins in the group had a coiled-coil domain of 21 amino acids or longer. This left 916 Caenorhabditis orthogroups including all SC proteins. We found that the median coiled-coil conservation score of SC proteins was significantly higher than that of the other coiled-coil proteins, confirming our comparisons to the entire proteome. We have included this analysis as a figure supplement to figure 2 (dot plot shown here and Figure 2 – figure supplement 1) and added text to the results and methods describing the analysis.
More broadly, this result suggests that our coiled-coil conservation score is more informative than a binary measure of coiled-coil domain prediction (i.e. presence/absence of coiled-coil). The additional information contained in the coiled-coil conservation score likely comes from the fact that we take into account whether or not the coiled-coil domains are aligned across species; which reflects a higher degree of secondary structure conservation. We believe that future work to develop better measures of conservation of secondary structures will hone our ability to identify conservation of other protein classes.
- We have clarified this point in our revised manuscript, highlighting that when analyzed as a group, indels are excluded in coiled-coils of Caenorhabditis SC proteins, and that significance is also observed for specific SC proteins where enough indels are present to perform statistical tests. Two of the SC proteins, SYP-2 and SYP-3, had only two indels each, preventing us from performing tests of significance. We have also added text to the discussion directly addressing the limitations of automatically-assigned gene annotations on the ability to test evolutionary pressures on indels genome-wide.
Reviewer #3 (Public Review):
The manuscript "Unconventional conservation reveals structure-function relationships in the synaptonemal complex" by Kursel, Cope, and Rog, describes a novel bioinformatics analysis of proteins in the eukaryotic synaptonemal complex (SC). The SC is a highly conserved structure that links paired homologs in prophase of meiosis, and in most organisms is required for the successful completion of interhomolog recombination. An enigmatic feature of SC proteins is that they are highly diverged between organisms, to the point where they are nearly unrecognizable by sequence alone except among closely related organisms. Kursel et al show that within the Caenorhabditis family of nematodes, SC proteins show a reproducible pattern of coiled-coil segments and highly conserved overall length, while their primary sequences are extremely diverged. They use these findings to develop a method to identify new SC candidate proteins in a diverged nematode, Pristionchus pacificus, and confirm that one of these candidates is the main SC transverse filament protein in this organism. Finally, the authors expand their analysis to SC proteins in flies (Drosophila melanogaster and relatives) and eutherian mammals, and show similar findings in these protein families. In the discussion, the authors describe an interesting and compelling theory that the coiled coils of SC proteins directly support phase separation/condensation of these proteins to aid assembly of the SC superstructure.
Overall, this work is well done, the findings are well-supported, and are of interest to meiosis researchers; especially those working directly on the SC. The manuscript is also well put-together: I could barely find a typo. From a broader perspective, however, I'm not convinced that the work provides a new paradigm for thinking about "conservation" in protein families and how to best detect it. Methods that use structural information to detect homology between highly diverged proteins beyond the capabilities of BLAST or even PSI-BLAST are well-developed (e.g. PHYRE2, HHPred, and others). The use of coiled-coil length as a metric for conservation, while it works nicely in the case of SC proteins, is likely to not be generalizable to other protein families. Even within SC proteins, the method does not seem to scale past specific families to, say, allow identification of homology between distantly-related eukaryotic groups (e.g. between Caenorhabditis and Drosophila or Caenorhabditis and eutherian mammals). To be fair, this failure to scale is not because of any limitation with the method; rather, simply that SC proteins diverge quickly through evolution. Overall, however, these limitations seem to limit the application of this method to the specialized case of SC proteins, thus limiting the audience and scope of the work.
We appreciate the reviewer’s consideration of possible limitations of our study. However, we disagree that this method, and the insights gained from it, will be limited to SC proteins. A clear demonstration is that the centrosomal protein SPD-5 (Centrosomin in Drosophila, CdkRap2 in mammals) cannot be identified across clades using sequence homology despite performing a conserved and fundamental cellular function. We hypothesize that similar forces have shaped the evolution of SPD-5 and other centrosomal proteins that are enriched in the same evolutionary class as SC proteins (Figure 3 – figure supplement 1). Functional tests of these predictions will be an exciting area of future research.
As this review notes, an exciting hypothesis stemming from our work is that proteins with diverged primary sequence and conserved secondary structures (coiled-coils, disordered protein domains or others) will be over-represented in condensates. Anecdotally this is indeed true, as both the SC and the centrosome were shown to be condensates. The burgeoning interest in condensates, and the development of tools to study them in vivo and in vitro, are bound to test the broad applicability of this hypothesis.
-

Evaluation Summary:
Although the synaptonemal complex (SC) is an essential, deeply conserved structure that holds meiotic chromosomes together, the constituent proteins evolve exceptionally rapidly. This rapid evolution in turn has hindered the identification of SC proteins based solely on sequence homology. This manuscript overcomes this challenge by developing and validating a clever protein structure-based approach that leverages sequence divergence - rather than sequence conservation - to identify novel SC components.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. The reviewers remained anonymous to the authors.)
-

Reviewer #1 (Public Review):
Kursel et al. examined the evolution of synaptonemal complex proteins in C.elegans. While the sequence of the SC proteins evolved rapidly analysis of the structure of SC central region proteins from Caenorhabditis, Drosophila and mammalian species revealed that the length and placement of the coiled-coil domains, as well as overall protein length, were highly conserved across species. This conservation in the structure of coiled-coil proteins within the SC led to the proposal that the conserved structural parameters of the SC proteins and their coiled-coil domains could be used to identify central region components of the SC in species where components could not be identified on sequence conservation alone. Kursel et al demonstrated their parameters could be used to identify a transverse filament protein of …
Reviewer #1 (Public Review):
Kursel et al. examined the evolution of synaptonemal complex proteins in C.elegans. While the sequence of the SC proteins evolved rapidly analysis of the structure of SC central region proteins from Caenorhabditis, Drosophila and mammalian species revealed that the length and placement of the coiled-coil domains, as well as overall protein length, were highly conserved across species. This conservation in the structure of coiled-coil proteins within the SC led to the proposal that the conserved structural parameters of the SC proteins and their coiled-coil domains could be used to identify central region components of the SC in species where components could not be identified on sequence conservation alone. Kursel et al demonstrated their parameters could be used to identify a transverse filament protein of the SC in the organism Pristionchus pacificus.
Due to high sequence divergence identifying SC proteins in new model systems has been challenging. The identification by Kursel et al. of potential search parameters to identify these diverged proteins will be useful to the those who work on the synaptonemal complex. This approach has the potential to applicable to other types of proteins that show rapid sequence divergence. As the mammalian, fly, and worm SC proteins all displayed different lengths and placements of their coiled-coil domains within their SC proteins this approach is limited by the availability of related identified sequences to the model organism of interest. Additionally, this approach may still yield multiple candidates that fit the structural parameters which will require additional means to ultimately identify the protein of interest. The data in the manuscript supports the authors' claims of structural conservation within SC proteins but only additional applications of their search methods will reveal how useful it is to search for other types of proteins based on structural features.
-

Reviewer #2 (Public Review):
In this article, Kursel and colleagues sought to identify evolutionary features of components of the SC the are evident in the absence of strict amino-acid conservation. After identifying three joint evolutionary properties of SC proteins - conservation of coiled-coil architecture, conservation of length and significant amino acid divergence - they show that these properties can be used to identify unknown SC proteins in divergent species. Overall, their general conclusion is very well supported and they do an excellent job functionally testing their approach by showing that one identified candidate for a novel SC protein in Pristionchus is in fact a component of the SC. In addition to providing new insight into the evolutionary forces that shape the evolution of SC proteins, this article provides new …
Reviewer #2 (Public Review):
In this article, Kursel and colleagues sought to identify evolutionary features of components of the SC the are evident in the absence of strict amino-acid conservation. After identifying three joint evolutionary properties of SC proteins - conservation of coiled-coil architecture, conservation of length and significant amino acid divergence - they show that these properties can be used to identify unknown SC proteins in divergent species. Overall, their general conclusion is very well supported and they do an excellent job functionally testing their approach by showing that one identified candidate for a novel SC protein in Pristionchus is in fact a component of the SC. In addition to providing new insight into the evolutionary forces that shape the evolution of SC proteins, this article provides new insight into how one might generally identify functionally similar or homologous proteins despite very deep divergence. Thus, this work has broader relevance to molecular evolution and evolution of protein structure.
There are some places where smaller conclusions need more support. In particular, it is not entirely clear that this triple pattern - conservation of coiled-coil architecture, conservation of length and significant amino acid divergence - is broadly applicable to SC components beyond Dipterans and Nematodes. In particular, the pattern is weaker in Eutherian mammals. Some further investigation is needed to claim that the pattern is similar in mammals. In addition, it is not clear if coiled-coil conservation rather than simply having a coiled-coil domain is important as a mark of SC proteins. A comparison of coiled-coil conservation among proteins that have coiled-coil domains would be needed for this conclusion. Finally, there should be some additional clarification that not all nematode SC proteins have a pattern of insertion and deletion that is limited to regions outside of the coil-coil domains.
-

Reviewer #3 (Public Review):
The manuscript "Unconventional conservation reveals structure-function relationships in the synaptonemal complex" by Kursel, Cope, and Rog, describes a novel bioinformatics analysis of proteins in the eukaryotic synaptonemal complex (SC). The SC is a highly conserved structure that links paired homologs in prophase of meiosis, and in most organisms is required for the successful completion of interhomolog recombination. An enigmatic feature of SC proteins is that they are highly diverged between organisms, to the point where they are nearly unrecognizable by sequence alone except among closely related organisms. Kursel et al show that within the Caenorhabditis family of nematodes, SC proteins show a reproducible pattern of coiled-coil segments and highly conserved overall length, while their primary …
Reviewer #3 (Public Review):
The manuscript "Unconventional conservation reveals structure-function relationships in the synaptonemal complex" by Kursel, Cope, and Rog, describes a novel bioinformatics analysis of proteins in the eukaryotic synaptonemal complex (SC). The SC is a highly conserved structure that links paired homologs in prophase of meiosis, and in most organisms is required for the successful completion of interhomolog recombination. An enigmatic feature of SC proteins is that they are highly diverged between organisms, to the point where they are nearly unrecognizable by sequence alone except among closely related organisms. Kursel et al show that within the Caenorhabditis family of nematodes, SC proteins show a reproducible pattern of coiled-coil segments and highly conserved overall length, while their primary sequences are extremely diverged. They use these findings to develop a method to identify new SC candidate proteins in a diverged nematode, Pristionchus pacificus, and confirm that one of these candidates is the main SC transverse filament protein in this organism. Finally, the authors expand their analysis to SC proteins in flies (Drosophila melanogaster and relatives) and eutherian mammals, and show similar findings in these protein families. In the discussion, the authors describe an interesting and compelling theory that the coiled coils of SC proteins directly support phase separation/condensation of these proteins to aid assembly of the SC superstructure.
Overall, this work is well done, the findings are well-supported, and are of interest to meiosis researchers; especially those working directly on the SC. The manuscript is also well put-together: I could barely find a typo. From a broader perspective, however, I'm not convinced that the work provides a new paradigm for thinking about "conservation" in protein families and how to best detect it. Methods that use structural information to detect homology between highly diverged proteins beyond the capabilities of BLAST or even PSI-BLAST are well-developed (e.g. PHYRE2, HHPred, and others). The use of coiled-coil length as a metric for conservation, while it works nicely in the case of SC proteins, is likely to not be generalizable to other protein families. Even within SC proteins, the method does not seem to scale past specific families to, say, allow identification of homology between distantly-related eukaryotic groups (e.g. between Caenorhabditis and Drosophila or Caenorhabditis and eutherian mammals). To be fair, this failure to scale is not because of any limitation with the method; rather, simply that SC proteins diverge quickly through evolution. Overall, however, these limitations seem to limit the application of this method to the specialized case of SC proteins, thus limiting the audience and scope of the work.
-