A novel immunopeptidomic-based pipeline for the generation of personalized oncolytic cancer vaccines
Curation statements for this article:-
Curated by eLife
Evaluation Summary:
This study describes an immunopeptidomic-based pipeline to discover new tumor antigens for the development of cancer vaccines. The pipeline is relatively straightforward and exploits molecular mimicry and tumor pathogen cross-reactive T-cells and would be interesting for cancer immunologists. If the utility of the pipeline were demonstrated in more diverse systems, including carcinogen-induced tumors and human settings, this work would provide an immediate impact to the immuno-oncology field and personalized cancer vaccine development.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #2 agreed to share their name with the authors.)
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Besides the isolation and identification of major histocompatibility complex I-restricted peptides from the surface of cancer cells, one of the challenges is eliciting an effective antitumor CD8+ T-cell-mediated response as part of therapeutic cancer vaccine. Therefore, the establishment of a solid pipeline for the downstream selection of clinically relevant peptides and the subsequent creation of therapeutic cancer vaccines are of utmost importance. Indeed, the use of peptides for eliciting specific antitumor adaptive immunity is hindered by two main limitations: the efficient selection of the most optimal candidate peptides and the use of a highly immunogenic platform to combine with the peptides to induce effective tumor-specific adaptive immune responses. Here, we describe for the first time a streamlined pipeline for the generation of personalized cancer vaccines starting from the isolation and selection of the most immunogenic peptide candidates expressed on the tumor cells and ending in the generation of efficient therapeutic oncolytic cancer vaccines. This immunopeptidomics-based pipeline was carefully validated in a murine colon tumor model CT26. Specifically, we used state-of-the-art immunoprecipitation and mass spectrometric methodologies to isolate >8000 peptide targets from the CT26 tumor cell line. The selection of the target candidates was then based on two separate approaches: RNAseq analysis and HEX software. The latter is a tool previously developed by Jacopo, 2020, able to identify tumor antigens similar to pathogen antigens in order to exploit molecular mimicry and tumor pathogen cross-reactive T cells in cancer vaccine development. The generated list of candidates (26 in total) was further tested in a functional characterization assay using interferon-γ enzyme-linked immunospot (ELISpot), reducing the number of candidates to six. These peptides were then tested in our previously described oncolytic cancer vaccine platform PeptiCRAd, a vaccine platform that combines an immunogenic oncolytic adenovirus (OAd) coated with tumor antigen peptides. In our work, PeptiCRAd was successfully used for the treatment of mice bearing CT26, controlling the primary malignant lesion and most importantly a secondary, nontreated, cancer lesion. These results confirmed the feasibility of applying the described pipeline for the selection of peptide candidates and generation of therapeutic oncolytic cancer vaccine, filling a gap in the field of cancer immunotherapy, and paving the way to translate our pipeline into human therapeutic approach.
Article activity feed
-

-

Author Response
Reviewer #1 (Public Review):
Identifying private peptides for generating personalised cancer vaccines is a promising approach to launch robust anti-tumor response; however, the challenges remain in developing an effective process to achieve that. In this manuscript, the authors present an interesting and powerful pipeline (PeptiCRAD) to achieve this goal by examining CT26 model. Overall, this manuscript is well written and presented. Despite that this work presents interesting findings and pipeline, I have the following concerns. I do feel that this manuscript will improve if these concerns can be addressed.
We thank the reviewer very much for having appreciated the quality and the originality of our work.
- It will be critical to confirm TILs and T cells in draining lymph nodes indeed recognise the peptide used in …
Author Response
Reviewer #1 (Public Review):
Identifying private peptides for generating personalised cancer vaccines is a promising approach to launch robust anti-tumor response; however, the challenges remain in developing an effective process to achieve that. In this manuscript, the authors present an interesting and powerful pipeline (PeptiCRAD) to achieve this goal by examining CT26 model. Overall, this manuscript is well written and presented. Despite that this work presents interesting findings and pipeline, I have the following concerns. I do feel that this manuscript will improve if these concerns can be addressed.
We thank the reviewer very much for having appreciated the quality and the originality of our work.
- It will be critical to confirm TILs and T cells in draining lymph nodes indeed recognise the peptide used in Figure 7-8 by ELISPOT of IFNg.
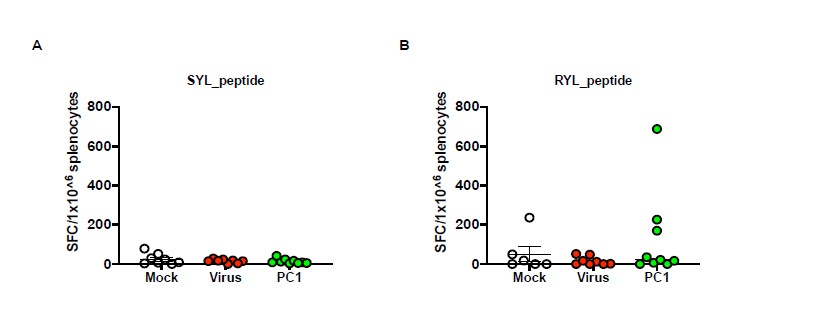
We agree with the reviewer´s comment as regard to confirming that TILS and T cells in draining lymph nodes recognise the peptide used in Figure 7-8 by functional characterization in an ELISPOT IFN-γ assay. In our experience, the ELISPOT assay works at the best when fresh samples are employed; additionally, the splenocytes are source of enough cells to test individual mouse reactivity to single peptide. To this end, as the samples from figures 7 and 8 were frozen, we decided to repeat the animal experiment according to figure 7 schedule treatment to perform then the ELISPOT on splenocytes freshly harvested from mice. Following the previous results, we selected the best group (PeptiCRAd1) to further investigate the peptide response; untreated mice (Mock) and Virus alone (VALO-mD901) were used as control as well. Interestingly, the peptide deconvolution showed T cell reactivity to one peptide (RYLPAPTAL, peptide 2) (Figure 1A) in the PeptiCRAd1 group, in contrast no T-cell reactivity was observed for SYLPPGTSL (peptide 1) (Figure 1B). These data highlighted the role of an individual antigen in eliciting specific anti-tumor T cell response, appearing an interested candidate for further proof of concept in animal experimental setting.
Figure 1 Interferon-γ Elispot results Harvested splenocytes from the treatment groups (as indicated in the figure) were functional characterized in an IFN-γ ELISPOT assay; individual response to SYLPPGTSL A) and RYLPAPTAL B) for each mouse is reported as IFN-γ spot forming cells (SFC)/106 splenocytes. The data are depicted as single dots plot and mean + SEM is shown. (Virus=VALO-mD901, PC=PeptiCRAd).
- It would be interesting to see if this pipeline can be used to identify human peptides in human melanomas.
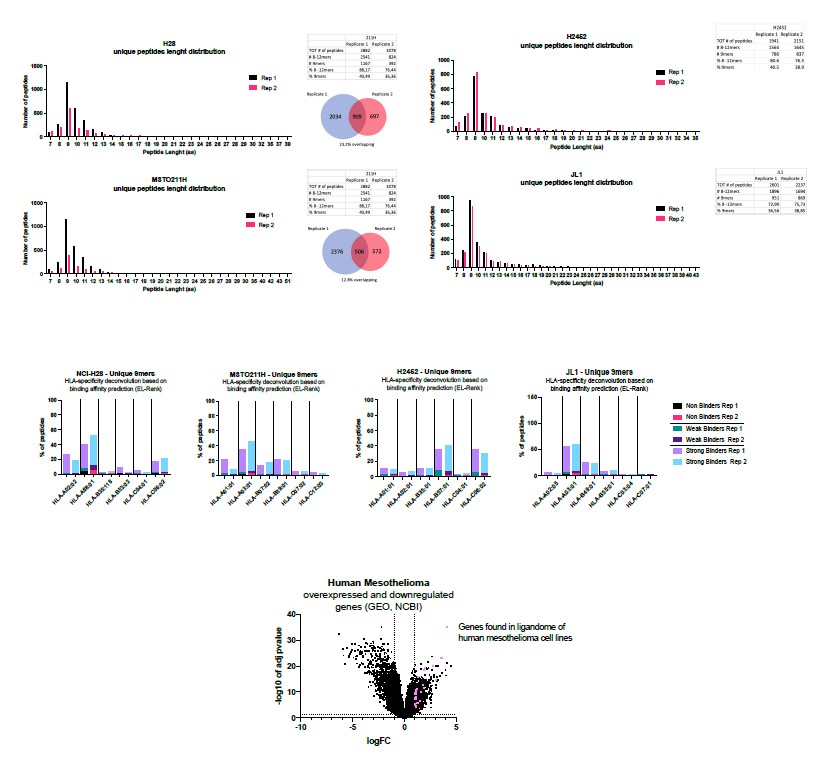
We thank the reviewer for pointing out that this pipeline can be used to identify human peptide in human melanomas. Indeed, the work here described is a proof concept meant to be translated in human setting. To this end, in the lab we have two projects on-going that are exploiting the same pipeline to investigate the human epithelioid and human mesothelioma ligandome landscape. Regarding this latter, we are investigating four different human cell lines (H2B, MSTO211H, H2452 and JL1). As shown in the picture below (Figure 2), the peptide length distribution showed an enrichment in 9mres in both replicates (Rep1 and Rep2), in line with a ligandome profile. The analysis of the binders revealed that most of them were good binders (according to EL-Rank score) for at least one of alleles for each cell line. Following the pipeline reported in this manuscript, to select candidate peptides we applied two different approaches; the first approach relied on RNA seq analysis to check which source proteins of the peptides isolated in the ligandome analysis were reported as upregulated or downregulated in resected tumor compared to normal tissues.
(Fromhttps://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE51024 GSE51024). The second approach (analysis still on-going) will be the use of HEX software.
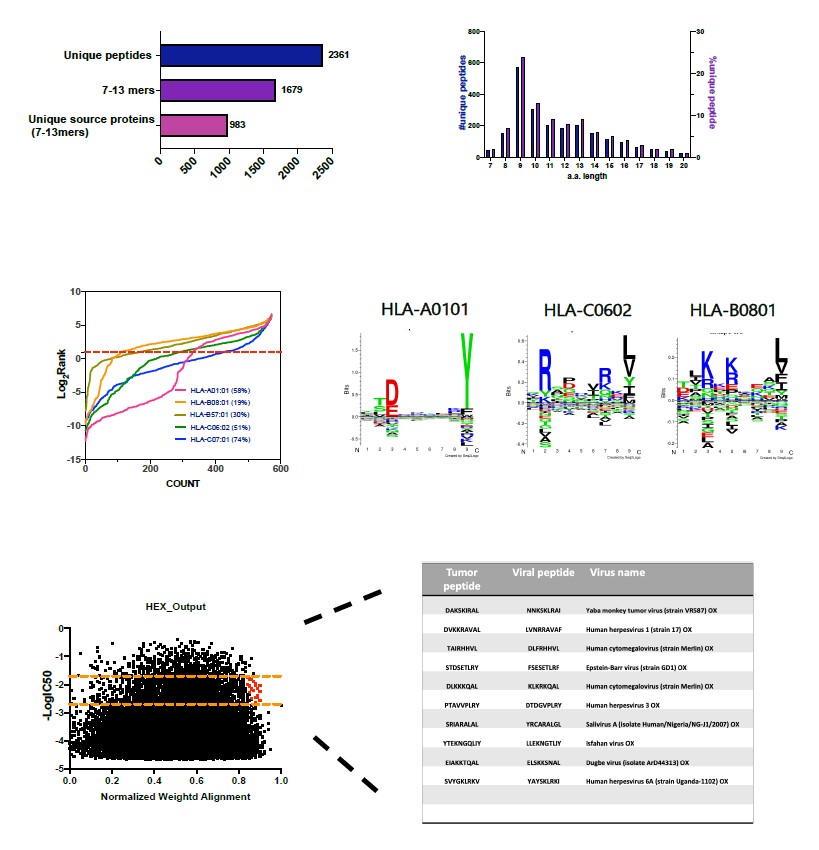
Regarding the epithelioid project, we have analysed the ligandome profile of two human cell lines: NEPS and VA-ES-BJ. Please find below the example of the ligandome analysis for VAES- BJ cell line (Figure 3). Overall, the analysis outcome was similar to published dataset (aminoacidic length distribution, Gibbs clustering profile, amount of binders) confirming the good quality of the ligandome landscape identified. Next, we applied HEX analysis to narrow down the list of peptide candidates for further test. We are currently in the stage of collecting more different epithelioid cell lines to expand our cohort of samples.
Figure 2 Mesothelioma project (data not published)
Figure 3 Epithelioid project (data not published)
-

Evaluation Summary:
This study describes an immunopeptidomic-based pipeline to discover new tumor antigens for the development of cancer vaccines. The pipeline is relatively straightforward and exploits molecular mimicry and tumor pathogen cross-reactive T-cells and would be interesting for cancer immunologists. If the utility of the pipeline were demonstrated in more diverse systems, including carcinogen-induced tumors and human settings, this work would provide an immediate impact to the immuno-oncology field and personalized cancer vaccine development.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #2 agreed to share their name with the authors.)
-

Reviewer #1 (Public Review):
Identifying private peptides for generating personalised cancer vaccines is a promising approach to launch robust anti-tumor response; however, the challenges remain in developing an effective process to achieve that. In this manuscript, the authors present an interesting and powerful pipeline (PeptiCRAD) to achieve this goal by examining CT26 model. Overall, this manuscript is well written and presented. Despite that this work presents interesting findings and pipeline, I have the following concerns. I do feel that this manuscript will improve if these concerns can be addressed.
1. It will be critical to confirm TILs and T cells in draining lymph nodes indeed recognise the peptide used in Figure 7-8 by ELISPOT of IFNg.
2. It would be interesting to see if this pipeline can be used to identify human peptides …
Reviewer #1 (Public Review):
Identifying private peptides for generating personalised cancer vaccines is a promising approach to launch robust anti-tumor response; however, the challenges remain in developing an effective process to achieve that. In this manuscript, the authors present an interesting and powerful pipeline (PeptiCRAD) to achieve this goal by examining CT26 model. Overall, this manuscript is well written and presented. Despite that this work presents interesting findings and pipeline, I have the following concerns. I do feel that this manuscript will improve if these concerns can be addressed.
1. It will be critical to confirm TILs and T cells in draining lymph nodes indeed recognise the peptide used in Figure 7-8 by ELISPOT of IFNg.
2. It would be interesting to see if this pipeline can be used to identify human peptides in human melanomas.
-

Reviewer #2 (Public Review):
Summary:
In Feola et al., the authors applied an immunopeptidomic-based pipeline to discover tumor antigens that can be incorporated into an adenovirus/vaccine platform to potentially cure Balb/c mice with established CT26 tumors. The authors performed immunopeptidomic experiments to isolate Kd/Dd-associated peptides, selected the most promising peptide candidates based on differential mRNA expression (mTEC and normal colon as control) and sequence similarities with pathogens (NEX software). Then, they tested a list of 26 peptide candidates in vitro (ELISPOT), selected the 6 most immunogenic peptides and tested their therapeutic potential in vivo using their PeptiCRAd vaccine platform in Balb/c mice with established CT26 tumors. Finally, they measured tumor growth and analyzed tumor infiltrating T cells in …
Reviewer #2 (Public Review):
Summary:
In Feola et al., the authors applied an immunopeptidomic-based pipeline to discover tumor antigens that can be incorporated into an adenovirus/vaccine platform to potentially cure Balb/c mice with established CT26 tumors. The authors performed immunopeptidomic experiments to isolate Kd/Dd-associated peptides, selected the most promising peptide candidates based on differential mRNA expression (mTEC and normal colon as control) and sequence similarities with pathogens (NEX software). Then, they tested a list of 26 peptide candidates in vitro (ELISPOT), selected the 6 most immunogenic peptides and tested their therapeutic potential in vivo using their PeptiCRAd vaccine platform in Balb/c mice with established CT26 tumors. Finally, they measured tumor growth and analyzed tumor infiltrating T cells in vaccinated mice bearing tumor cells to confirm the feasibility of applying their pipeline for the generation of therapeutic oncolytic cancer vaccine.
Strengths of the methods and results:
The pipeline appears to be straightforward. A clear strength of the study is that the authors applied immunopeptidomics to measure MHCI-peptides that are genuinely presented on the cell surface. The method provides physical measurements of MHCI-peptides using mass spectrometry. This is a direct approach that differ from the application of MHC-peptide binding prediction algorithms, which are cheap/highly accessible and useful but often lead to relatively large numbers of false positive peptides that are non-immunogenic and therapeutically irrelevant. Another strength is that the authors tested their peptide candidates in vivo (in mice) using their PeptiCRAd vaccine platform.
Weaknesses of the methods and results:
The study provides a complete pipeline, from antigen discovery to pre-clinical validation in mice using a vaccine platform. This could be viewed as a strength, but there are important limitations in each component of the pipeline, making the overall pipeline relatively suboptimal. The rational for the selection of the peptide candidates could be better explained and additional controls could be used throughout the study. To what extent the selected tumor antigens are tumor-specific is unclear. The therapeutical potential of HEX-identified peptides compared to previously reported TSA (mutated or aberrantly expressed) is not evaluated in this study. Most importantly, the anti-tumor response measured in vivo appear to be MHCIpeptide-independent since the uncoated adenovirus (VALO-mD901) improved tumor growth control in the infection site, raising doubts about the overall approach. No conceptual advancement is provided by the study.
Is the aim achieved, and results support their conclusions?
There is a lot of work in this study. The authors aimed to show the feasibility of applying their pipeline to identify tumor antigens, and then test the therapeutic potential of those peptides in mice; this is achieved. However, it would be important to not oversell the pipeline since many tumor antigens that have been identified are non-immunogenic and have little impact in vivo. The current pipeline is a good start but still need significant improvements to rationalize the peptide selection process and to clearly understand why those peptides are, or are not, immunogenetic in vivo. This is important as the authors envision the development of a robust platform that can be successful in the clinic.
Impact and significance of the work:
The work shows that immunopeptidomics is useful to identify tumor peptide antigens. The peptide selection is also based on viral molecular mimicry (using the HEX software), which is relatively new in the field and certainly worth exploring by others. I envision that this type of pipeline can be useful in the context of a biotech company that aims to rapidly identify and test potential peptide targets.
-

Reviewer #3 (Public Review):
The goal of the study was to establish a first-in-the-world personalized oncolytic viral vaccine pipeline that covers the entire process, from the identification of candidate vaccine targets from the primary tumor to the in vivo efficacy testing.
By using their pipeline, the authors identified peptides that could be successfully loaded onto the oncolytic virus. In addition, the generated oncolytic vaccine was effective not only in the treated tumor but also in the untreated distant tumor. These observations indeed demonstrate that their pipeline platform can successfully generate oncolytic viral vaccines that can induce a strong systemic anti-tumor immunity.
However, this study's major weakness is that it has no experimental data that support their conclusion that this pipeline can be translationally applied …
Reviewer #3 (Public Review):
The goal of the study was to establish a first-in-the-world personalized oncolytic viral vaccine pipeline that covers the entire process, from the identification of candidate vaccine targets from the primary tumor to the in vivo efficacy testing.
By using their pipeline, the authors identified peptides that could be successfully loaded onto the oncolytic virus. In addition, the generated oncolytic vaccine was effective not only in the treated tumor but also in the untreated distant tumor. These observations indeed demonstrate that their pipeline platform can successfully generate oncolytic viral vaccines that can induce a strong systemic anti-tumor immunity.
However, this study's major weakness is that it has no experimental data that support their conclusion that this pipeline can be translationally applied to the clinical setting as "PERSONALIZED" vaccine development.
Personalized medicine, by definition, is a treatment that is tailored specifically to a particular patient. One of the challenges in developing effective personalized medicine for cancer patients is how to utilize the materials taken from the patients to generate the effective treatment in a timely manner so that it can be therapeutically utilized before the patients reach the untreatable stage of the disease.
While the pipeline demonstrated in the study was effective, the authors had access to the primary tumor material, an immortalized transplantable tumor cell line grown in vitro, to identify the peptide ahead of time, then to treat cancer developed by injecting the same cell line into the mice. Although this perfectly validates the pipeline as a capable system to induce anti-tumor immunity, it fails to demonstrate the challenges of applying this as personalized medicine in the future, as they concluded.
-