Volume growth in animal cells is cell cycle dependent and shows additive fluctuations
Curation statements for this article:-
Curated by eLife
Evaluation Summary:
The regulation of cell growth is crucial for our understanding of how cells control their size as well as how they balance cell mass and volume. While recent studies carefully measured single cell mass trajectories during the cell cycle, revealing complex growth patterns, the volume growth patterns of animal cells are poorly understood. In this interesting study, Cadart et al. now present high-precision measurements of 1700 HeLa cell growth trajectories and offering evidence for the mechanisms that regulate volume growth-rate fluctuations. This is an important demonstration of cell autonomous volume regulation.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #2 agreed to share their name with the authors.)
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
The way proliferating animal cells coordinate the growth of their mass, volume, and other relevant size parameters is a long-standing question in biology. Studies focusing on cell mass have identified patterns of mass growth as a function of time and cell cycle phase, but little is known about volume growth. To address this question, we improved our fluorescence exclusion method of volume measurement (FXm) and obtained 1700 single-cell volume growth trajectories of HeLa cells. We find that, during most of the cell cycle, volume growth is close to exponential and proceeds at a higher rate in S-G2 than in G1. Comparing the data with a mathematical model, we establish that the cell-to-cell variability in volume growth arises from constant-amplitude fluctuations in volume steps rather than fluctuations of the underlying specific growth rate. We hypothesize that such ‘additive noise’ could emerge from the processes that regulate volume adaptation to biophysical cues, such as tension or osmotic pressure.
Article activity feed
-

-

Author Response:
Reviewer #1:
The dependence of cell volume growth rate on cell size and cell cycle is a long-standing fundamental question that has traditionally been addressed by using unicellular model organisms with simple geometry, for which rough volume estimates can be obtained from bright field images. While it became soon apparent that the volume growth rate depends on cell volume, the experimental error associated with such measurements made it difficult to determine the exact dependencies. This challenge is even more significant for animal cells, whose complex and dynamic geometry makes accurate volume measurements extremely difficult. Other measures for cell size, including mass or fluorescent reporters for protein content, partially bypassed this problem. However, it becomes increasingly clear that cell mass and volume …
Author Response:
Reviewer #1:
The dependence of cell volume growth rate on cell size and cell cycle is a long-standing fundamental question that has traditionally been addressed by using unicellular model organisms with simple geometry, for which rough volume estimates can be obtained from bright field images. While it became soon apparent that the volume growth rate depends on cell volume, the experimental error associated with such measurements made it difficult to determine the exact dependencies. This challenge is even more significant for animal cells, whose complex and dynamic geometry makes accurate volume measurements extremely difficult. Other measures for cell size, including mass or fluorescent reporters for protein content, partially bypassed this problem. However, it becomes increasingly clear that cell mass and volume are not strictly coupled, making accurate volume measurements essential. In their previous work, Cadart and colleagues established a 'fluorescent exclusion method', which allows accurate volume measurements of cells with complex geometry. In the present manuscript, Cadart et al. now take the next step and measure the growth trajectories of 1700 HeLa cell cycles with further improved accuracy, providing new insights into animal cell growth.
They convincingly demonstrate that throughout large parts of the cell cycle, individual cells exhibit exponential growth, with the volume-normalized specific growth rate moderately increasing after G1-phase. At the very early stages of the cell cycle, cells exhibit a more complex growth behavior. The authors then go on and analyze the growth rate fluctuations of individual cells, identifying a decrease of the variance of the specific growth rate with cell volume and observed time scale. The authors conclude that the observed growth fluctuations are consistent with additive noise of the absolute growth rate.
The experiments and analysis presented by Cadart et al. are carefully and well executed, and the insights provided (as well as the method established) are an important contribution to our understanding of cell growth. My major concern is that the observed fluctuation pattern seems largely consistent with what would be expected if the fluctuations stem from experimental measurement noise. This fact is appropriately acknowledged, and the authors aim to address this issue by analyzing background noise. However, further controls may be necessary to unambiguously attribute the measured noise to biological fluctuations, rather than experimental error.
We thank the reviewer for their positive feedback and for the appreciation of our work. We performed a series of experimental controls to address the main issue regarding the measured fluctuation pattern, which indicate that it should be of biological origin.
1.) To address whether the observed fluctuations could be due to experimental error, the authors analyze the fluctuations recorded in a cell-sized area of the background, and find that the background fluctuations are small compared to the fluctuations of the volume measurements. I think this is a very important control that supports the interpretation of the authors. However, I am not convinced that the actual measurement error is necessarily of the same amplitude as the fluctuations of the background. The background control will control for example for variations of light intensity and fluctuations of the fluorophore intensity. But what about errors in the cell segmentation? Or movement of the cells in 3D, which could be relevant because the collected light might be dependent on the distance from the surface? Is cell autofluorescence relevant at all? I am aware that accurately estimating the experimental error is exceptionally difficult, and I am also not entirely sure what would be the perfect control (if it even exists). Nevertheless, I think more potential sources of error should be addressed before the measured noise can be confidently attributed to biological sources. Maybe the authors could measure objects with constant volume over time, for example vesicles? As long as the segmented area contains the complete cell, the measured volume should not change if the area is increased. Is this the case?
We are grateful to the reviewer for all these useful suggestions. We performed all these controls on the sources of noise, and we discuss them in the revised manuscript.
2.) I am particularly puzzled by the fact that even at the timescale of the frame rate, fluctuations seem not to be correlated between 2 consecutive time points (Fig. 5-S2b). This seems plausible for (some) sources of experimental error. Maybe an experiment with fast time resolution would reveal the timescale over which the fluctuations persist - which could then give us a hint about the source?
We performed this analysis, finding an autocorrelation time of a few minutes, and we report our results below:
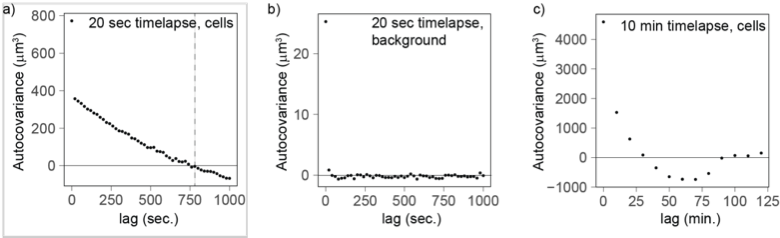
In the main text and in the new Figure 5 – Supplement 3, we report the results of newly performed 20 sec timelapse experiments over one hour to investigate the timescale of volume fluctuations. The autocvariance function analysis on the detrended curves shows that fluctuations decay over a few minutes (Figure 5 – Supplement 3a-c), a timescale that matches the analysis of the 10 min timelapse experiments.
Copy of Figure 5 – Supplement 3: Autocovariance analysis shows that the timescale of volume fluctuation is around 760 seconds. a) Cells measured every 20 sec (n=177) and linearly detrended reach a covariance of 0 at a lag of 760 sec. b) As a control, the background fluctuations are not autocorrelated (20 sec, n=92), providing further evidence that cell volume fluctuations likely have biological origin. c) The autocovariance analysis for cells measured every 10 min confirms that fluctuations covary for a lag of 10-20 min.
3.) The authors use automated smoothing of the measurement and removed outliers based on an IQR-criteria. While this seems reasonable if the aim is to get a robust measurement of the average behavior, I find it questionable with respect to the noise measurements. Since no minimum time scale has been associated with the fluctuations interpreted as biological in origin, what is the justification of removing 'outliers', i.e. the feature that the authors are actually interested in? Why would the largest fluctuations be of technical origin, and the smaller fluctuations exclusively biological?
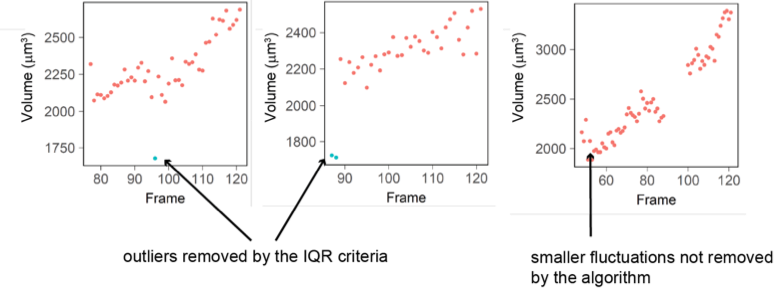
The IQR-criteria is designed to remove only rare and obvious outliers (i.e. a jump in volume of more than 15% in 1 frame -10 minutes- which arguably cannot happen biologically). Fluctuations of smaller range are kept (see examples below). We looked back at the raw data and calculated that the IQR filtering removes a total of 337 measurement points out of 99935 initial points (0.03% of the points).
Figure D: Three examples of single cell trajectories with raw volume measurement (red dots) and points removed with the IQR filtering (blue dots). The IQR criteria is very stringent and removes only the very large ‘bumps’ in cell volume measured (2 left plots) while it keeps fluctuations of smaller amplitude (right plot).
4.) If I understood correctly, each volume trajectory spans one complete cell cycle. If this is the case, does Fig. 1e imply that many cell cycles take less than 2-3 hours? Is this really the case, and if so, what are the implications for some of the interpretations (especially the early cell cycle part)?
In this study, we performed experiments on a time scale comparable to the cell cycle time (~ 24hours) and recorded single-cell volume trajectories. Since the cells are not synchronized, we have very few complete cell cycles (~ 100, Fig. 1f). Fig. 1e shows the distribution of the duration of all individual curves, regardless of the fraction of the cell cycle they span, hence the very short duration for some cells.
Reviewer #2:
In this paper, the authors use a volume exclusion-based measurements to quantify single cell trajectories of volume increase in HeLa cells. The study represents one of the most careful measurements on volume regulation in animal cells and presents evidence for feedback mechanisms that slow the growth of larger cells. This is an important demonstration of cell autonomous volume regulation.
While the subject matter of the present study is important, the insights provided are significantly limited because the authors did not place their findings in the context of previous literature. The authors present what seems to be remarkably accurate single cell growth trajectories. In animal cells, a joint dependency of growth rate on cell size and cell cycle stage has been previously reported (see Elife 2018 PMID: 29889021 and Science 2009 PMID: 19589995). In Ginzberg et al, it is reported "Our data revealed that, twice during the cell cycle, growth rates are selectively increased in small cells and reduced in large cells". Nonetheless, these previous studies do not negate the novelty in Cadart et al. While both Cadart and Ginzberg investigate a dependency of cellular growth rate on cell size and cell cycle stage, the two studies are complimentary. This is because, while Ginzberg characterise the growth in cell mass, Cadart characterise the growth in cell volume. The authors should compare the findings from these previous studies with their own and draw conclusions from the similarities and differences. Are the cell cycle stage dependent growth rate similar or different when cell size is quantified as mass or volume? Does the faster growth of smaller cells (the negative correlation of growth rate and cell size) occur in different cell cycle stages when growth is quantified by volume as compared to mass?
We are grateful to the reviewer for their appreciation of the value of our study. Following their remarks, we have extended our Discussion section to incorporate a more careful discussion of these findings. We believe that the main contribution of our study is finding evidence of phase- dependent regulation of growth rate and identifying an additive noise on volume steps, this noise has constant amplitude, hence fluctuations of specific growth rate decrease with volume, but specific growth rate (in the bulk of the cell cycle) does not decrease.
-

Evaluation Summary:
The regulation of cell growth is crucial for our understanding of how cells control their size as well as how they balance cell mass and volume. While recent studies carefully measured single cell mass trajectories during the cell cycle, revealing complex growth patterns, the volume growth patterns of animal cells are poorly understood. In this interesting study, Cadart et al. now present high-precision measurements of 1700 HeLa cell growth trajectories and offering evidence for the mechanisms that regulate volume growth-rate fluctuations. This is an important demonstration of cell autonomous volume regulation.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #2 agreed to share …
Evaluation Summary:
The regulation of cell growth is crucial for our understanding of how cells control their size as well as how they balance cell mass and volume. While recent studies carefully measured single cell mass trajectories during the cell cycle, revealing complex growth patterns, the volume growth patterns of animal cells are poorly understood. In this interesting study, Cadart et al. now present high-precision measurements of 1700 HeLa cell growth trajectories and offering evidence for the mechanisms that regulate volume growth-rate fluctuations. This is an important demonstration of cell autonomous volume regulation.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #2 agreed to share their name with the authors.)
-

Reviewer #1 (Public Review):
The dependence of cell volume growth rate on cell size and cell cycle is a long-standing fundamental question that has traditionally been addressed by using unicellular model organisms with simple geometry, for which rough volume estimates can be obtained from bright field images. While it became soon apparent that the volume growth rate depends on cell volume, the experimental error associated with such measurements made it difficult to determine the exact dependencies. This challenge is even more significant for animal cells, whose complex and dynamic geometry makes accurate volume measurements extremely difficult. Other measures for cell size, including mass or fluorescent reporters for protein content, partially bypassed this problem. However, it becomes increasingly clear that cell mass and volume are …
Reviewer #1 (Public Review):
The dependence of cell volume growth rate on cell size and cell cycle is a long-standing fundamental question that has traditionally been addressed by using unicellular model organisms with simple geometry, for which rough volume estimates can be obtained from bright field images. While it became soon apparent that the volume growth rate depends on cell volume, the experimental error associated with such measurements made it difficult to determine the exact dependencies. This challenge is even more significant for animal cells, whose complex and dynamic geometry makes accurate volume measurements extremely difficult. Other measures for cell size, including mass or fluorescent reporters for protein content, partially bypassed this problem. However, it becomes increasingly clear that cell mass and volume are not strictly coupled, making accurate volume measurements essential. In their previous work, Cadart and colleagues established a 'fluorescent exclusion method', which allows accurate volume measurements of cells with complex geometry. In the present manuscript, Cadart et al. now take the next step and measure the growth trajectories of 1700 HeLa cell cycles with further improved accuracy, providing new insights into animal cell growth.
They convincingly demonstrate that throughout large parts of the cell cycle, individual cells exhibit exponential growth, with the volume-normalized specific growth rate moderately increasing after G1-phase. At the very early stages of the cell cycle, cells exhibit a more complex growth behavior. The authors then go on and analyze the growth rate fluctuations of individual cells, identifying a decrease of the variance of the specific growth rate with cell volume and observed time scale. The authors conclude that the observed growth fluctuations are consistent with additive noise of the absolute growth rate.
The experiments and analysis presented by Cadart et al. are carefully and well executed, and the insights provided (as well as the method established) are an important contribution to our understanding of cell growth. My major concern is that the observed fluctuation pattern seems largely consistent with what would be expected if the fluctuations stem from experimental measurement noise. This fact is appropriately acknowledged, and the authors aim to address this issue by analyzing background noise. However, further controls may be necessary to unambiguously attribute the measured noise to biological fluctuations, rather than experimental error.
Major points:
1.) To address whether the observed fluctuations could be due to experimental error, the authors analyze the fluctuations recorded in a cell-sized area of the background, and find that the background fluctuations are small compared to the fluctuations of the volume measurements. I think this is a very important control that supports the interpretation of the authors. However, I am not convinced that the actual measurement error is necessarily of the same amplitude as the fluctuations of the background. The background control will control for example for variations of light intensity and fluctuations of the fluorophore intensity. But what about errors in the cell segmentation? Or movement of the cells in 3D, which could be relevant because the collected light might be dependent on the distance from the surface? Is cell autofluorescence relevant at all? I am aware that accurately estimating the experimental error is exceptionally difficult, and I am also not entirely sure what would be the perfect control (if it even exists). Nevertheless, I think more potential sources of error should be addressed before the measured noise can be confidently attributed to biological sources. Maybe the authors could measure objects with constant volume over time, for example vesicles? As long as the segmented area contains the complete cell, the measured volume should not change if the area is increased. Is this the case?
2.) I am particularly puzzled by the fact that even at the timescale of the frame rate, fluctuations seem not to be correlated between 2 consecutive time points (Fig. 5-S2b). This seems plausible for (some) sources of experimental error. Maybe an experiment with fast time resolution would reveal the timescale over which the fluctuations persist - which could then give us a hint about the source?
3.) The authors use automated smoothing of the measurement and removed outliers based on an IQR-criteria. While this seems reasonable if the aim is to get a robust measurement of the average behavior, I find it questionable with respect to the noise measurements. Since no minimum time scale has been associated with the fluctuations interpreted as biological in origin, what is the justification of removing 'outliers', i.e. the feature that the authors are actually interested in? Why would the largest fluctuations be of technical origin, and the smaller fluctuations exclusively biological?
4.) If I understood correctly, each volume trajectory spans one complete cell cycle. If this is the case, does Fig. 1e imply that many cell cycles take less than 2-3 hours? Is this really the case, and if so, what are the implications for some of the interpretations (especially the early cell cycle part)?
-

Reviewer #2 (Public Review):
In this paper, the authors use a volume exclusion-based measurements to quantify single cell trajectories of volume increase in HeLa cells. The study represents one of the most careful measurements on volume regulation in animal cells and presents evidence for feedback mechanisms that slow the growth of larger cells. This is an important demonstration of cell autonomous volume regulation.
While the subject matter of the present study is important, the insights provided are significantly limited because the authors did not place their findings in the context of previous literature. The authors present what seems to be remarkably accurate single cell growth trajectories. In animal cells, a joint dependency of growth rate on cell size and cell cycle stage has been previously reported (see Elife 2018 PMID: …
Reviewer #2 (Public Review):
In this paper, the authors use a volume exclusion-based measurements to quantify single cell trajectories of volume increase in HeLa cells. The study represents one of the most careful measurements on volume regulation in animal cells and presents evidence for feedback mechanisms that slow the growth of larger cells. This is an important demonstration of cell autonomous volume regulation.
While the subject matter of the present study is important, the insights provided are significantly limited because the authors did not place their findings in the context of previous literature. The authors present what seems to be remarkably accurate single cell growth trajectories. In animal cells, a joint dependency of growth rate on cell size and cell cycle stage has been previously reported (see Elife 2018 PMID: 29889021 and Science 2009 PMID: 19589995). In Ginzberg et al, it is reported "Our data revealed that, twice during the cell cycle, growth rates are selectively increased in small cells and reduced in large cells". Nonetheless, these previous studies do not negate the novelty in Cadart et al. While both Cadart and Ginzberg investigate a dependency of cellular growth rate on cell size and cell cycle stage, the two studies are complimentary. This is because, while Ginzberg characterise the growth in cell mass, Cadart characterise the growth in cell volume. The authors should compare the findings from these previous studies with their own and draw conclusions from the similarities and differences. Are the cell cycle stage dependent growth rate similar or different when cell size is quantified as mass or volume? Does the faster growth of smaller cells (the negative correlation of growth rate and cell size) occur in different cell cycle stages when growth is quantified by volume as compared to mass?
-