Haploinsufficiency of the essential gene Rps12 causes defects in erythropoiesis and hematopoietic stem cell maintenance
Curation statements for this article:-
Curated by eLife
eLife assessment
This study will be of interest to scientists within the field of hematopoiesis and ribosome biology. The paper provides evidence that haploinsufficiency of the mouse ribosomal protein gene Rps12 results in a number of phenotypes including defects in the production of specific blood cells and loss of hematopoietic stem cell quiescence. This work adds to the growing body of evidence that specific cell populations are particularly sensitive to disruption of mRNA translation machinery.
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Ribosomal protein (Rp) gene haploinsufficiency can result in Diamond-Blackfan Anemia (DBA), characterized by defective erythropoiesis and skeletal defects. Some mouse Rp mutations recapitulate DBA phenotypes, although others lack erythropoietic or skeletal defects. We generated a conditional knockout mouse to partially delete Rps1 2. Homozygous Rps12 deletion resulted in embryonic lethality. Mice inheriting the Rps12 KO/+ genotype had growth and morphological defects, pancytopenia, and impaired erythropoiesis. A striking reduction in hematopoietic stem cells (HSCs) and progenitors in the bone marrow (BM) was associated with decreased ability to repopulate the blood system after competitive and non-competitive BM transplantation. Rps12 KO/+ lost HSC quiescence, experienced ERK and MTOR activation, and increased global translation in HSC and progenitors. Post-natal heterozygous deletion of Rps12 in hematopoietic cells using Tal1-Cre-ERT also resulted in pancytopenia with decreased HSC numbers. However, post-natal Cre-ERT induction led to reduced translation in HSCs and progenitors, suggesting that this is the most direct consequence of Rps12 haploinsufficiency in hematopoietic cells. Thus, RpS12 has a strong requirement in HSC function, in addition to erythropoiesis.
Article activity feed
-

-

Author Response
Reviewer #3 (Public Review):
In this manuscript, the authors studied the erythropoiesis and hematopoietic stem/progenitor cell (HSPC) phenotypes in a ribosome gene Rps12 mutant mouse model. They found that RpS12 is required for both steady and stress hematopoiesis. Mechanistically, RpS12+/- HSCs/MPPs exhibited increased cycling, loss of quiescence, protein translation rate, and apoptosis rates, which may be attributed to ERK and Akt/mTOR hyperactivation. Overall, this is a new mouse model that sheds light into our understanding of Rps gene function in murine hematopoiesis. The phenotypic and functional analysis of the mice are largely properly controlled, robust, and analyzed.
A major weakness of this work is its descriptive nature, without a clear mechanism that explains the phenotypes observed in RpS12+/- mice. It …
Author Response
Reviewer #3 (Public Review):
In this manuscript, the authors studied the erythropoiesis and hematopoietic stem/progenitor cell (HSPC) phenotypes in a ribosome gene Rps12 mutant mouse model. They found that RpS12 is required for both steady and stress hematopoiesis. Mechanistically, RpS12+/- HSCs/MPPs exhibited increased cycling, loss of quiescence, protein translation rate, and apoptosis rates, which may be attributed to ERK and Akt/mTOR hyperactivation. Overall, this is a new mouse model that sheds light into our understanding of Rps gene function in murine hematopoiesis. The phenotypic and functional analysis of the mice are largely properly controlled, robust, and analyzed.
A major weakness of this work is its descriptive nature, without a clear mechanism that explains the phenotypes observed in RpS12+/- mice. It is possible that the counterintuitive activation of ERK/mTOR pathway and increased protein synthesis rate is a compensatory negative feedback. Direct mechanism of Rps12 loss could be studied by ths acute loss of Rps12, which is doable using their floxed mice. At the minimum, this can be done in mammalian hematopoietic cell lines.
We thank the reviewer for pointing this out. We have addressed this question by developing a new inducible conditional knockout Rps12 mouse model (see response below to major point 1).
Below are some specific concerns need to be addressed.
- Line 226. The authors conclude that "Together, these results suggest that RpS12 plays an essential role in HSC function, including self-renewal and differentiation." The reviewer has three concerns regarding this conclusion and corresponding Figure3. 1) The data shows that RpS12+/- mice have decreased number of both total BM cells and multiple subpopulations of HSPCs. The frequency of HSPC subpopulations should also be shown to clarify if the decreased HSPC numbers arises from decreased total BM cellularity or proportionally decrease in frequency. 2) This figure characterizes phenotypic HSPC in BM by flow and lineage cells in PB by CBC. HSC function and differentiation are not really examined in this figure, except for the colony assay in Figure 3K. BMT data in Figure4 is actually for HSC function and differentiation. So the conclusion here should be rephrased. 3) Since all LT-, ST-HSCs, as well as all MPPs are decreased in number, how can the authors conclude that Rps12 is important for HSC differentiation? No experiments presented here were specifically designed to address HSC differentiation.
We thank the reviewer for this excellent point. We think that the main defect is in HSC and progenitor maintenance, rather than in HSC differentiation. This is consistent with the decrease in multiple HSC and progenitor populations, as observed both by calculating absolute numbers and by frequency of the parent population (see new Supplementary Figures S2C-S2C). We have removed any references to altered differentiation from the text.
We added data on the population frequency in the Supplementary Figure 2. And in the corresponding text. See lines 221-235.
- Figure 3A and 5E. The flow cytometry gating of HSC/MPP is not well performed or presented, especially HSC plot. Populations are not well separated by phenotypic markers. This concerns the validity of the quantification data.
We chose a better representative HSC plot and included it in the Figure 3A
- It is very difficult to read bone marrow cytospin images in Fig 6F without annotation of cell types shown in the figure. It appears that WT and +/- looked remarkably different in terms of cell size and cell types. This mouse may have other profound phenotypes that need detailed examination, such as lineage cells in the BM and spleen, and colony assays for different types of progenitors, etc.
The purpose of the bone marrow cytospin images in Figure 6F was to show the high number of apoptotic cells in the bone marrow of Rps12 KO/+ mice compared with controls. The differences in apoptosis in the LSK and myeloid progenitor populations are quantified in the flow cytometry data shown in Figure 6G-H. A detailed quantitative analysis of different bone marrow cell populations and their relative frequencies is also shown in Figures 2 and 3. In Rps12 KO/+ bone marrow, we observed a significant decrease in multiple stem cell and progenitor populations.
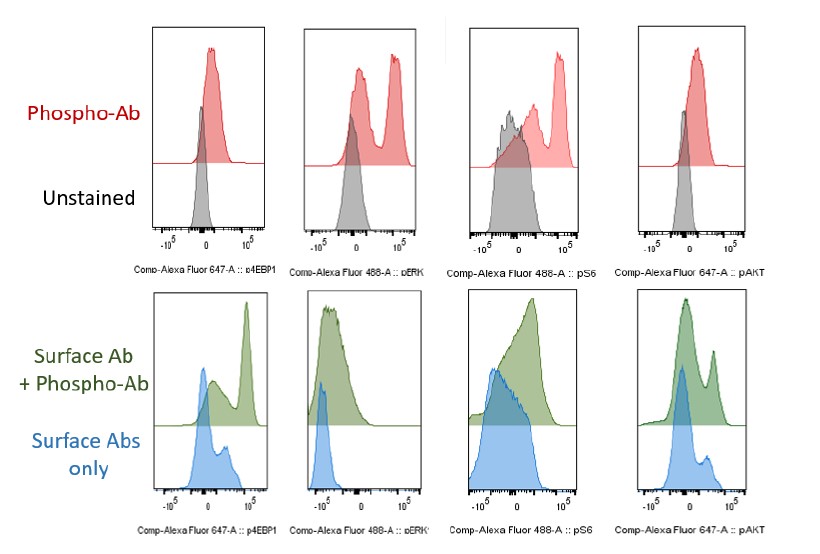
- For all the intracellular phospho-flow shown in Fig7, both a negative control of a fluorescent 2nd antibody only and a positive stimulus should be included. It is very concerning that no significant changes of pAKT and pERK signaling (MFI) after SCF stimulation from the histogram in WT LSKs. There are no distinct peaks that indicate non-phospho-proteins and phosphoproteins. This casts doubt on the validity of results. It is possible though that Rsp12+/- have very high basal level of activation of pAKT/mTOR and pERK pathway. This again may point to a negative feedback mechanism of Rps12 haploinsufficiency.
It is true that we did not observe an increase in pAKT, p4EBP1, or pERK in control cells in every case. This is often an issue with these specific phospho-flow cytometry antibodies, as they are not very sensitive, and the response to SCF is very time-dependent. We did observe an increase in pS6 with SCF in both LSK cells and progenitors (Figure 7B, E). However, the main point of this experiment was to assess the basal level of signaling in Rps12 KO/+ vs control cells. We did not observe hypersensitivity of RpS12 cells to SCF, but we did observe significant increases in pAKT, pS6, p4EBP1, and pERK in Rsp12 KO/+ LSK cells.
To address the concern about the validity of staining, please see the requested flow histograms for unstained vs individual Phospho-antibodies (Ab): p4EBP1, pERK, pS6 and pAKT (Figure R1 for reviewers) below. Additionally, since staining with the surface antibodies potentially can change the peak, we are including additional an control of the cell surface antibodies vs full sample with surface antibodies and Phospho-Ab: p4EBP1, pERK, pS6 and pAKT. We can include this figure in the Supplementary Data if requested.
- The authors performed in vitro OP-Puro assay to assess the global protein translation in different HSPC subpopulations. 1) Can the authors provide more information about the incubation media, any cytokine or serum included? The incubation media with supplements may boost the overall translation status, although cells from WT and RpS12+/- are cultured side by side. Based on this, in vivo OP-Puro assay should be performed in both genotypes. 2) Polysome profiling assay should be performed in primary HSPCs, or at least in hematopoietic cell lines. It is plausible that RpS12 haploinsufficiency may affect the content of translational polysome fractions.
We are including these details in the methods section: for in vitro OP-Puro assay (lines 555565) cells were resuspended in DMEM (Corning 10-013-CV) media supplemented with 50 µM β-mercaptoethanol (Sigma) and 20 µM OPP (Thermo Scientific C10456). Cells were incubated for 45 minutes at 37°C and then washed with Ca2+ and Mg2+ free PBS. No additional cytokines were added.
We did not perform polysome profiles. Polysome profiling of mutant stem and progenitor cells would be very challenging, as their numbers are much reduced. We now deem this of reduced interest, given the conclusion of the revised manuscript that RpS12 haploinsufficiency reduces overall translation. Also, because in RpS12-floxed/+;SCL-CRE-ERT mouse model with acute deletion of RpS12 we observed the expected decrease in translation in HSCs using the same ex vivo OPP protocol, we did not follow up with in vivo OPP treatment,
-

eLife assessment
This study will be of interest to scientists within the field of hematopoiesis and ribosome biology. The paper provides evidence that haploinsufficiency of the mouse ribosomal protein gene Rps12 results in a number of phenotypes including defects in the production of specific blood cells and loss of hematopoietic stem cell quiescence. This work adds to the growing body of evidence that specific cell populations are particularly sensitive to disruption of mRNA translation machinery.
-

Reviewer #1 (Public Review):
In humans, mutations in specific ribosomal protein genes and ribosome assembly factors cause a group of diseases collectively known as ribosomopathies. Patients with these diseases typically display a number of remarkably similar tissue specific phenotypes including anemia and craniofacial abnormalities. The causes of the tissue specificity of these disorders have long remained an outstanding question in the field, and more recent evidence points to the induction of nucleolar stress which triggers a p53-dependent response and cell death. In previous work, the authors have shown that loss and gain of Drosophila Rps12 causes a number of unexpected phenotypes. This current paper seeks to investigate the function of Rps12 in mice.
The authors generate a conditional knockout allele within the mouse Rps12 locus …
Reviewer #1 (Public Review):
In humans, mutations in specific ribosomal protein genes and ribosome assembly factors cause a group of diseases collectively known as ribosomopathies. Patients with these diseases typically display a number of remarkably similar tissue specific phenotypes including anemia and craniofacial abnormalities. The causes of the tissue specificity of these disorders have long remained an outstanding question in the field, and more recent evidence points to the induction of nucleolar stress which triggers a p53-dependent response and cell death. In previous work, the authors have shown that loss and gain of Drosophila Rps12 causes a number of unexpected phenotypes. This current paper seeks to investigate the function of Rps12 in mice.
The authors generate a conditional knockout allele within the mouse Rps12 locus and show that homozygous loss of Rps12 results in early embryonic lethality, while heterozygous mutants display a number of cell specific defects in the hematopoietic system. The authors provide evidence that haploinsufficiency of Rps12 results in erythropoiesis defects that worsen with age, a decrease in the number of hematopoietic progenitor cells, and disruption of hematopoietic stem cell (HSC) quiescence correlated with a failure of mutant HSCs to reconstitute peripheral blood. Strikingly, loss of Rps12 results in increased translation in HSCs and early progenitors, marked by activation of MEK/ERK and ARK/TOR signaling pathways.
Strengths
The paper provides new evidence that loss of Rps12 results in a number of specific defects in the hematopoietic system. The phenotypic characterization is rigorous and clearly described in the text. The observations Rps12 heterozygotes exhibit increases in protein synthesis and loss of HSC quiescence are interesting and warrant further investigation. This paper will have broad appeal to those interested in development, stem cell maintenance, ribosome biology, and ribosomopathies.Weaknesses
The Rps12 gene has two embedded snoRNAs, the disruption of which could contribute to all of the described phenotypes. Additional work is needed to confirm that the mutant phenotypes are caused specifically by loss of Rps12 -

Reviewer #2 (Public Review):
Previous work from the authors' lab has shown that the classical 'Minute' phenotypes in Drosophila depend on the ribosomal protein Rps12, suggesting that Rps12 is a sensor of deficits in other ribosomal proteins (Rp). Increasing the dose of Rps12 enhances 'Minute' phenotypes, while loss of Rps12 suppresses them. However, Rps12+/- heterozygous flies do not display 'Minute' phenotypes.
In the current manuscript, the authors examine the consequences of deleting Rps12 in mice to explore its potential role in translational regulation and hematopoiesis. Homozygosity for an Rps12 null mutation is embryonic lethal, while heterozygous Rps12+/- mutant mice exhibit defects in growth, skeletal abnormalities, hydrocephalus and stroke. Consistent with other mouse Rp mutants, Rps12+/- mutant mice have a block in erythroid …
Reviewer #2 (Public Review):
Previous work from the authors' lab has shown that the classical 'Minute' phenotypes in Drosophila depend on the ribosomal protein Rps12, suggesting that Rps12 is a sensor of deficits in other ribosomal proteins (Rp). Increasing the dose of Rps12 enhances 'Minute' phenotypes, while loss of Rps12 suppresses them. However, Rps12+/- heterozygous flies do not display 'Minute' phenotypes.
In the current manuscript, the authors examine the consequences of deleting Rps12 in mice to explore its potential role in translational regulation and hematopoiesis. Homozygosity for an Rps12 null mutation is embryonic lethal, while heterozygous Rps12+/- mutant mice exhibit defects in growth, skeletal abnormalities, hydrocephalus and stroke. Consistent with other mouse Rp mutants, Rps12+/- mutant mice have a block in erythroid maturation and reduced spleen size. Hematopoietic stem and progenitor cell (HSPC) numbers are reduced in the bone marrow and are defective in repopulation transplant assays. Unexpectedly, Rps12+/- mutants show loss of HSC quiescence associated with AKT/MTOR and ERK pathway activation and increased global translation, a phenomenon that has not previously been reported in other Rp mutants. The authors conclude that Rps12 is critical for the maintenance of HSC quiescence and function.
Strengths
The data reported in this manuscript nicely complement the existing literature on the functional effects of Rp mutations in mammalian hematopoiesis and development with loss of HSC quiescence and increased global translation in the Rps12 deficient mice. These unexpected findings will be of broad interest to scientists working in the field of ribosome assembly, ribosomopathies and hematopoiesis.Weaknesses
It remains unclear mechanistically how Rps12 haploinsufficiency activates the AKT/MTOR and ERK signaling pathways. It is also unclear to what extent the reported phenotypes might be indirect consequences of perturbing the expression of two small nucleolar RNA genes that are present in Rps12 introns 4 and 5 or a consequence of TP53 activation, which is known to influence the phenotype in other examples of Rp deletion mouse models. To fully justify the conclusions that the authors wish to draw, it would be important to assess the effect of the heterozygous Rps12+/- mutation on Rps12 protein expression, ribosomal subunit assembly and rRNA processing. -

Reviewer #3 (Public Review):
In this manuscript, the authors studied the erythropoiesis and hematopoietic stem/progenitor cell (HSPC) phenotypes in a ribosome gene Rps12 mutant mouse model. They found that RpS12 is required for both steady and stress hematopoiesis. Mechanistically, RpS12+/- HSCs/MPPs exhibited increased cycling, loss of quiescence, protein translation rate, and apoptosis rates, which may be attributed to ERK and Akt/mTOR hyperactivation. Overall, this is a new mouse model that sheds light into our understanding of Rps gene function in murine hematopoiesis. The phenotypic and functional analysis of the mice are largely properly controlled, robust, and analyzed.
A major weakness of this work is its descriptive nature, without a clear mechanism that explains the phenotypes observed in RpS12+/- mice. It is possible that the …
Reviewer #3 (Public Review):
In this manuscript, the authors studied the erythropoiesis and hematopoietic stem/progenitor cell (HSPC) phenotypes in a ribosome gene Rps12 mutant mouse model. They found that RpS12 is required for both steady and stress hematopoiesis. Mechanistically, RpS12+/- HSCs/MPPs exhibited increased cycling, loss of quiescence, protein translation rate, and apoptosis rates, which may be attributed to ERK and Akt/mTOR hyperactivation. Overall, this is a new mouse model that sheds light into our understanding of Rps gene function in murine hematopoiesis. The phenotypic and functional analysis of the mice are largely properly controlled, robust, and analyzed.
A major weakness of this work is its descriptive nature, without a clear mechanism that explains the phenotypes observed in RpS12+/- mice. It is possible that the counterintuitive activation of ERK/mTOR pathway and increased protein synthesis rate is a compensatory negative feedback. Direct mechanism of Rps12 loss could be studied by ths acute loss of Rps12, which is doable using their floxed mice. At the minimum, this can be done in mammalian hematopoietic cell lines.
Below are some specific concerns need to be addressed.
1. Line 226. The authors conclude that "Together, these results suggest that RpS12 plays an essential role in HSC function, including self-renewal and differentiation." The reviewer has three concerns regarding this conclusion and corresponding Figure3. 1) The data shows that RpS12+/- mice have decreased number of both total BM cells and multiple subpopulations of HSPCs. The frequency of HSPC subpopulations should also be shown to clarify if the decreased HSPC numbers arises from decreased total BM cellularity or proportionally decrease in frequency. 2) This figure characterizes phenotypic HSPC in BM by flow and lineage cells in PB by CBC. HSC function and differentiation are not really examined in this figure, except for the colony assay in Figure 3K. BMT data in Figure4 is actually for HSC function and differentiation. So the conclusion here should be rephrased. 3) Since all LT-, ST-HSCs, as well as all MPPs are decreased in number, how can the authors conclude that Rps12 is important for HSC differentiation? No experiments presented here were specifically designed to address HSC differentiation.
2. Figure 3A and 5E. The flow cytometry gating of HSC/MPP is not well performed or presented, especially HSC plot. Populations are not well separated by phenotypic markers. This concerns the validity of the quantification data.
3. It is very difficult to read bone marrow cytospin images in Fig 6F without annotation of cell types shown in the figure. It appears that WT and +/- looked remarkably different in terms of cell size and cell types. This mouse may have other profound phenotypes that need detailed examination, such as lineage cells in the BM and spleen, and colony assays for different types of progenitors, etc.
4. For all the intracellular phospho-flow shown in Fig7, both a negative control of a fluorescent 2nd antibody only and a positive stimulus should be included. It is very concerning that no significant changes of pAKT and pERK signaling (MFI) after SCF stimulation from the histogram in WT LSKs. There are no distinct peaks that indicate non-phospho-proteins and phospho-proteins. This casts doubt on the validity of results. It is possible though that Rsp12+/- have very high basal level of activation of pAKT/mTOR and pERK pathway. This again may point to a negative feedback mechanism of Rps12 haploinsufficiency.
5. The authors performed in vitro OP-Puro assay to assess the global protein translation in different HSPC subpopulations. 1) Can the authors provide more information about the incubation media, any cytokine or serum included? The incubation media with supplements may boost the overall translation status, although cells from WT and RpS12+/- are cultured side by side. Based on this, in vivo OP-Puro assay should be performed in both genotypes. 2) Polysome profiling assay should be performed in primary HSPCs, or at least in hematopoietic cell lines. It is plausible that RpS12 haploinsufficiency may affect the content of translational polysome fractions.
-