Disintegration promotes protospacer integration by the Cas1-Cas2 complex
Curation statements for this article:-
Curated by eLife
Evaluation Summary:
In this manuscript, an in vitro Cas1-Cas2 model system is used to study the reaction used to insert foreign DNA elements into a CRISPR array during the adaptive phase of immunity. The authors propose that hydrolysis of one end of the transposon DNA may be the primary mechanism for the insertion of very small DNA elements (which are difficult to bend tightly) that are found for the proto spacer sequences, and that cellular repair pathways are responsible for ligating the CRISPR array back together in vivo. The findings additionally suggest that water-mediated disintegration has an unappreciated role in the generation of CRISPR arrays as part of the bacterial immune response. These hypotheses are intriguing and of potential interest to those in the CRISPR field. However, it is unclear how this in vitro study, which does not monitor the full the reaction (directionality is lost due to the lack of a PAM sequence in the substrate and several required cellular factors are missing), relates to transposition as it occurs in vivo. Overall, this is an interesting study that challenges the current thinking in the field, but it does not present sufficient evidence to establish the physiological significance of the observed effects, thereby limiting its potential broader impact.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. The reviewers remained anonymous to the authors.)
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
‘Disintegration’—the reversal of transposon DNA integration at a target site—is regarded as an abortive off-pathway reaction. Here, we challenge this view with a biochemical investigation of the mechanism of protospacer insertion, which is mechanistically analogous to DNA transposition, by the Streptococcus pyogenes Cas1-Cas2 complex. In supercoiled target sites, the predominant outcome is the disintegration of one-ended insertions that fail to complete the second integration event. In linear target sites, one-ended insertions far outnumber complete protospacer insertions. The second insertion event is most often accompanied by the disintegration of the first, mediated either by the 3′-hydroxyl exposed during integration or by water. One-ended integration intermediates may mature into complete spacer insertions via DNA repair pathways that are also involved in transposon mobility. We propose that disintegration-promoted integration is functionally important in the adaptive phase of CRISPR-mediated bacterial immunity, and perhaps in other analogous transposition reactions.
Article activity feed
-

Author Response:
We have now revised the manuscript to address the helpful comments and criticisms from the reviewers. The revised manuscript includes additional experiments demonstrating that inclusion of Csn2/Cas9 in the in vitro assays does not suppress the disintegration activity of Cas1-Cas2 to favor integration. These additional factors do not confer strand selectivity on integration either. Furthermore, the results of integration reactions using substrates mimicking PAM-containing pre- spacers have also been added.
New figures and figure modifications at a glance:
- The new Figure 2 shows Cas1-Cas2 reactions in a linear target site and the effects of Csn2 and/or Cas9 on proto-spacer insertion into this target (Reviewer 1).
The original Figure 2 (with slight modifications) is now moved to ’Supplementary Data’ as Figure 2-figure …
Author Response:
We have now revised the manuscript to address the helpful comments and criticisms from the reviewers. The revised manuscript includes additional experiments demonstrating that inclusion of Csn2/Cas9 in the in vitro assays does not suppress the disintegration activity of Cas1-Cas2 to favor integration. These additional factors do not confer strand selectivity on integration either. Furthermore, the results of integration reactions using substrates mimicking PAM-containing pre- spacers have also been added.
New figures and figure modifications at a glance:
- The new Figure 2 shows Cas1-Cas2 reactions in a linear target site and the effects of Csn2 and/or Cas9 on proto-spacer insertion into this target (Reviewer 1).
The original Figure 2 (with slight modifications) is now moved to ’Supplementary Data’ as Figure 2-figure supplement 2, and shows proto-spacer insertion by Cas1-Cas2 into a nicked linear target site (Reviewer 2). Figure 2 is the only one in the main set of figures that has been extensively modified.
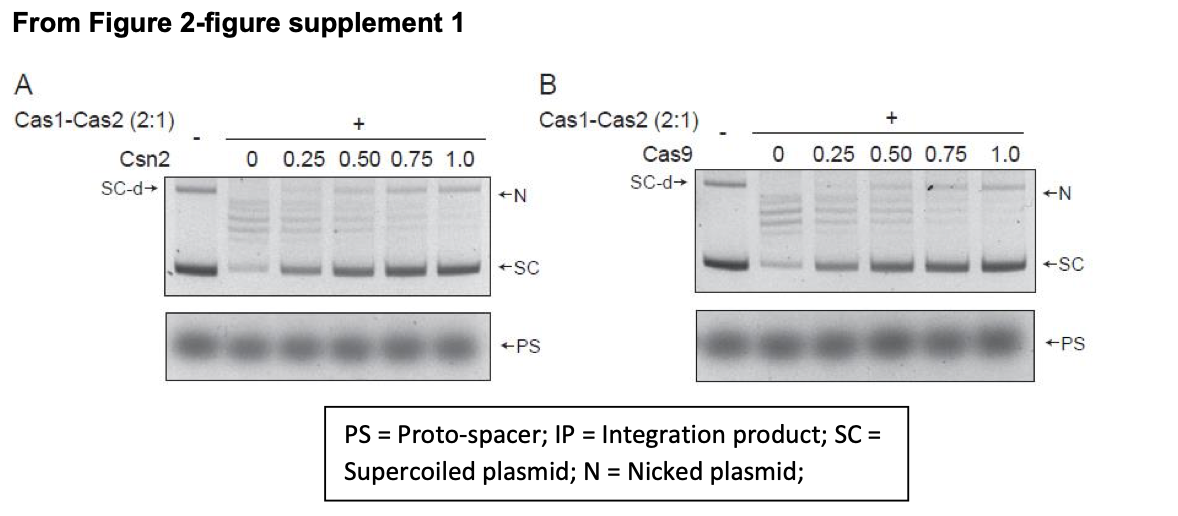
The new Figure 2-figure supplement 1 (under ‘Supplementary Data’) shows the effects of Csn2, Cas9 or both on proto-spacer integration-disintegration by Cas1-Cas2 when the target site is present in a supercoiled plasmid (Reviewer 1).
The new Figure 4-figure supplement 1 lists the sequences of the full- and half-target sites used for the reactions shown in Figure 4 (Reviewer 2).
The new Figure 2-figure supplement 3 shows the insertion properties of PAM-containing pre- spacer mimics in reactions with Cas1-Cas2 alone or supplemented with Csn2, Cas9 or both (Reviewer 1).
The new Figure 6-figure supplement 1 gives a structural perspective of the trombone substrates used for the reactions shown in Figure 6B, C (Reviewer 1).
The original Supplementary Figure S8 showing assays for PAM-specific cleavage by Cas1- Cas2 has been removed (Reviewer 1).
There are no changes in the other figures under ‘Supplementary Data’, although several have new numbers consistent with the revisions made.
Public Review (Reviewers #1 and #2):
The present work is a critical extension of the in vitro biochemical activities of the Cas1- Cas2 complex described by Wright and Doudna (Nat Struct Mol Biol, 2016; 23: 876-883). We have kept all experimental conditions nearly identical to those used by these authors to make the results from the two studies directly comparable. Importantly, we now show that the prior model for proto-spacer integration into the CRISPR locus by Cas1-Cas2 is an oversimplification of a much more nuanced mechanism.
While both reviewers recognize the importance of our findings in challenging the current thinking on the adaptation mechanism of CRISPR immunity, they express reservations as to whether the in vitro results recapitulate the in vivo mechanism of spacer acquisition. This seems to us to be too broad a criticism from which few (if any) biochemical experiments can be immune.
Our key finding is that disintegration during the second step of proto-spacer integration generates a DNA structure that has all the hallmarks of a DNA damage intermediate that the bacterial repair machinery can readily process into an authentic integration product. We invoke no new or ad hoc mechanisms, and the model we propose fits neatly into the DNA gap-filling mechanisms known to operate in DNA transposition pathways.
The proto-spacer is functionally a ‘micro-transposon’, whose shortness imposes severe torsional strain on the transposition intermediate that precedes the final integration product. In vitro experiments suggest that transcription is potentially capable of resolving this intermediate (Budhathoki et al., Nat Struct Mol Biol, 2020, 27: 489-99). In principle, replication can also accomplish this task. Our study now demonstrates that simply nicking the DNA (disintegration) is an equally effective solution for relieving the topological stress accompanying integration. DNA loose ends can then be readily tied up by the bacterial repair machinery.
We concur with the concluding sentence of reviewer 2, “The simple conclusion that Cas1- Cas2 catalyzed hydrolysis of a phosphodiester may relieve strain and allow productive transposition to occur doesn’t get emphasized enough in my opinion.” We have now expanded on this point in the revised ‘Discussion’.
Reviewer #1:
In addition, the in vitro system used here is only partially reconstituted. The substrates lack a PAM sequence, which is necessary for protospacers to be incorporated in the correct orientation and may help direct the first integration event to the L-R junction. Presumably because of this all the reactions presented do not analyze the orientation of the incorporated prespacer sequence. Cas9 and Csn2 are also absent (as are other potentially required host factors), which are necessary for correct integration in vivo.
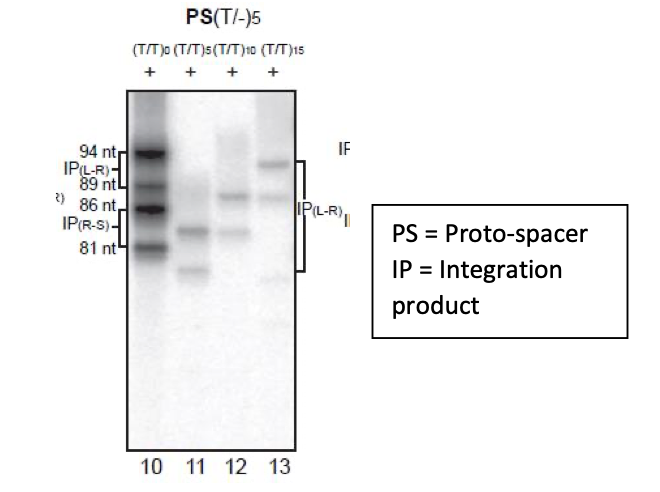
1A. Strand specificity: The in vitro integration reactions with the Cas1-Cas2 complex were done using a protospacer of the optimal size (26 nt on each strand with the four 3’- proximal bases on each strand as unpaired). Either proto-spacer strand is equally competent to initiate the strand transfer reaction, as could be inferred from Figure 3 of the original submission. Here, reactions utilized modified proto-spacers that differed in their top and bottom strand lengths. They gave two insertion products (IP) each at the L-R (leader-repeat) and R-S (repeat-spacer) junctions of a normal target site. In modified targets in which integration was limited to just the L- R junction, two insertion products were formed. One panel of Figure 3 (which is retained in the revised manuscript) showing the four insertion products from the normal target (lane 10) and two from the modified targets (lanes 11-13) for a protospacer with 26 nt and 31 nt long strands is displayed below.
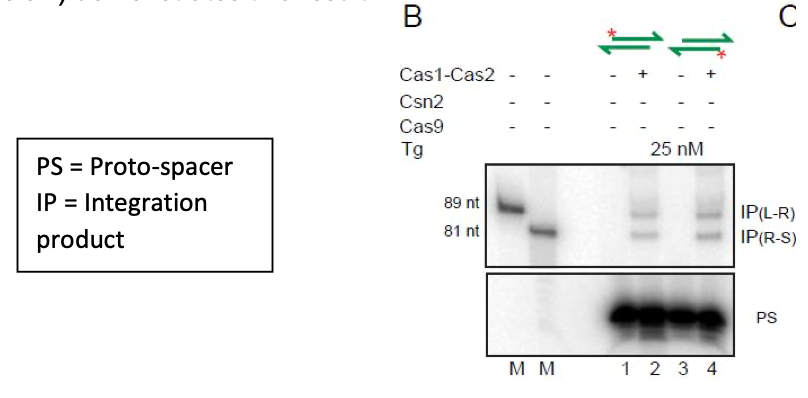
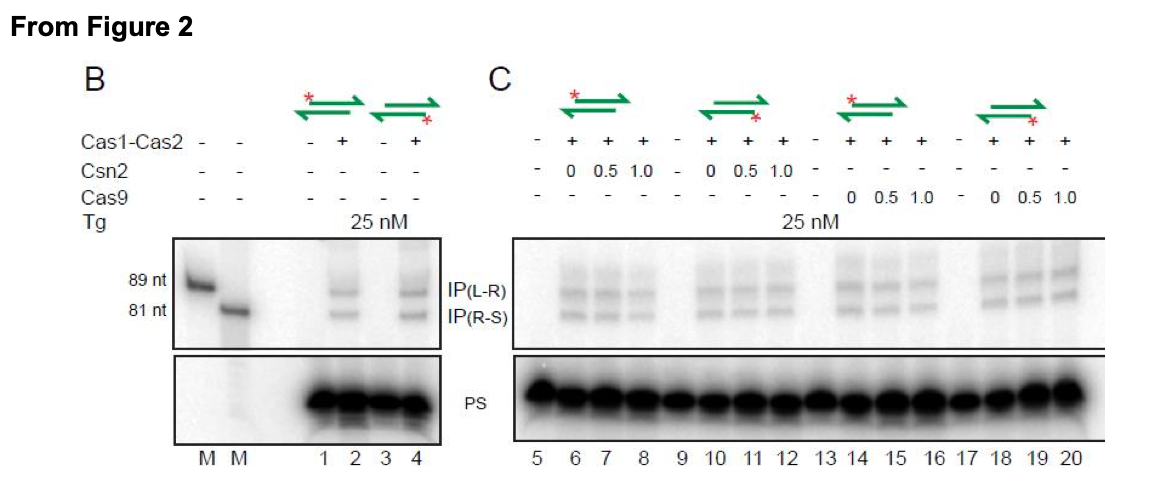
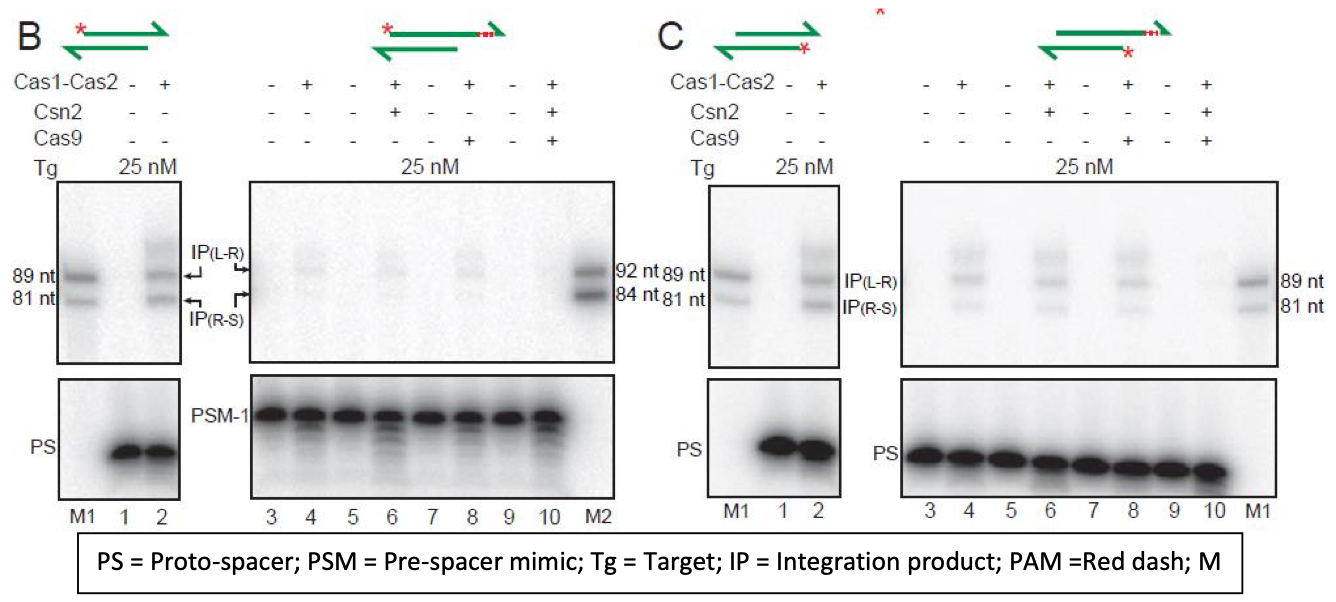
The ability of either proto-spacer strand to initiate integration is now more directly shown in Figure 2 (new) of the revised manuscript. Here the labeled top or bottom strand of the proto- spacer (PS) gave insertion products (IP) at the L-R and R-S junctions of the target site. Panel B of Figure 2 (pasted below) demonstrates this result.
1B. Cas9, Csn2 included reactions: The data for reactions containing Csn2 or Cas9 or both were not shown previously, as they did not alter Cas1-Cas2 activity by promoting strand specificity of integration or suppressing disintegration. These results are now shown in the revised Figure 2 (linear target) and the new Figure 2-figure supplement 1 (supercoiled target). Portions of these figures are shown below.
The relevant revised text describing the lack of strand specificity to proto-spacer integration by Cas1-Cas2 and the Csn2/Cas9 effects on integration is pasted below.
Page 15, lines 229-235.
"Unlike orientation-specific proto-spacer integration in vivo, Cas1- Cas2 reactions in vitro showed no strand-specificity (Figure 2B). This bias-free insertion of the top or bottom strand from the proto-spacer was unchanged by the addition of Csn2 or Cas9 or both to the reactions (Figure 2C-E). These proteins, singly or in combiantion, also failed to stabilize proto-spacer integrations in the supercoiled plasmid target (Figure 2-figure supplement 1). Instead, they inhibited plasmid relaxation. Inhibition could occur at the level of integration per se or strand rotation during integration-disintegration"
1C. PAM-containing substrates: We have now tested Cas1-Cas2 activity (with and without added Csn2 or Cas9 or both) on PAM-containing substrates that mimic ‘pre-spacers’, Figure 2- figure supplement 3 (new).
In these substrates, a proto-spacer strand of the standard length (26 nt; lacking PAM or its complement) is inserted at the L-R junction with higher efficiency than the longer strand (containing PAM or its complement). Following the first integration at L-R, the pre-spacer mimics containing > 26 nt in one strand or both strands are inhibited in the second strand transfer to the R-S junction. A portion of Figure 2-figure supplement 3 illustrating theses points is shown below.
The revised ‘Results’ section has the following added description of the activities of PAM- containing pre-spacer mimics.
Pages 16-19, lines 265-297. Cas1-Cas2 activity on pre-spacer mimics carrying the PAM sequence
"The strand cleavage and strand transfer steps of proto-spacer insertion at the CRISPR locus must engender safeguards against self-targeting of the inserted spacer as well as its non-functional orientation. However, no strand selectivity is seen in the in vitro Cas1-Cas2 reactions with already processed proto-spacers lacking the PAM sequence (Figures 2 and 3). By coordinating PAM- specific cleavage of a pre-spacer with transfer of this cleaved strand to the L-R junction, the inserted spacer will be in the correct orientation to generate a functional crRNA. To examine this possibility, we tested the integration characteristics of pre-spacer mimics containing the PAM sequence.
The inclusion of PAM or PAM and its complement in the integration substrates (Figure 2- figure supplement 3A) did not confer strand specificity on reactions with Cas1-Cas2 alone or with added Csn2, Cas9 or both (Figure 2-figure supplement 3B-E). Optimal integration by Cas1-Cas2 occurred with the 26 nt strands of the native protospacer with their 4 nt 3’-overhangs (Figure 2- figure supplement 3B-E; lanes 2). The pre-spacer mimics containing one or both > 26 nt strands had reduced integration competence (Figure 2-figure supplement 3B-E; lanes 4). Even here, the 26 nt strand with the 4 nt overhang (Figure 2-figure supplement 3C; lane 4) was preferred in integration over the longer 29nt PAM-containing strand (Figure 2-figure supplement 3D; lane 4) or the 33 nt PAM complement-containing strand (Figure 2-figure supplement 3E; lane 4). In contrast to the processed proto-spacer that gave nearly equal integration at L-R and R-S, IP(L- R) ≈ IP(R-S) (Figure 2-figure supplement 3B-E; lanes 2), the longer pre-spacer mimics were inhibited in integration at R-S, IP(L-R) > IP(R-S) (Figure 2-figure supplement 3B-E lanes 4). This is the expected outcome if the initial strand transfer occurs at L-R, and a ruler-like mechanism orients the reactive 3’-hydroxyl for the second strand transfer at R-S. This sequential two-step scheme for proto-spacer integration is consistent with the results shown in Figure 3 as well. These reaction features were not modulated by Csn2 or Cas9 (Figure 2-figure supplement 3B-E; lanes 6 and 8), although Csn2 plus Cas9 was inhibitory (Figure 2-figure supplement 3B-E; lanes 10).
There is no evidence for integration accompanying PAM-specific cleavage in our in vitro reactions. In the E. coli CRISPR system, Cas1-Cas2 is apparently sufficient for PAM-specific cleavage in vitro (22). By contrast, in the S. pyogenes system, cleavage is attributed to Cas9 or as yet uncharacterized bacterial nuclease(s) (35). The mechanism for generating an integration- proficient and orientation-specific proto-spacer, which may not be conserved among CRISPR systems, is poorly understood at this time."
-

Evaluation Summary:
In this manuscript, an in vitro Cas1-Cas2 model system is used to study the reaction used to insert foreign DNA elements into a CRISPR array during the adaptive phase of immunity. The authors propose that hydrolysis of one end of the transposon DNA may be the primary mechanism for the insertion of very small DNA elements (which are difficult to bend tightly) that are found for the proto spacer sequences, and that cellular repair pathways are responsible for ligating the CRISPR array back together in vivo. The findings additionally suggest that water-mediated disintegration has an unappreciated role in the generation of CRISPR arrays as part of the bacterial immune response. These hypotheses are intriguing and of potential interest to those in the CRISPR field. However, it is unclear how this in vitro study, which …
Evaluation Summary:
In this manuscript, an in vitro Cas1-Cas2 model system is used to study the reaction used to insert foreign DNA elements into a CRISPR array during the adaptive phase of immunity. The authors propose that hydrolysis of one end of the transposon DNA may be the primary mechanism for the insertion of very small DNA elements (which are difficult to bend tightly) that are found for the proto spacer sequences, and that cellular repair pathways are responsible for ligating the CRISPR array back together in vivo. The findings additionally suggest that water-mediated disintegration has an unappreciated role in the generation of CRISPR arrays as part of the bacterial immune response. These hypotheses are intriguing and of potential interest to those in the CRISPR field. However, it is unclear how this in vitro study, which does not monitor the full the reaction (directionality is lost due to the lack of a PAM sequence in the substrate and several required cellular factors are missing), relates to transposition as it occurs in vivo. Overall, this is an interesting study that challenges the current thinking in the field, but it does not present sufficient evidence to establish the physiological significance of the observed effects, thereby limiting its potential broader impact.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. The reviewers remained anonymous to the authors.)
-

Reviewer #1 (Public Review):
In this manuscript Ma and colleagues present a biochemical investigation into integration by the Spy Cas1-Cas2 integrase, a key component of the type II CRISPR immune response. From this work they challenge the view that disintegration (the reverse of integration) only functions as an off-pathway reaction (proof reading) and propose that disintegration-promoted integration is functionally important for integration into CRISPR arrays.
CRISPR integration proceeds through two steps. In the first, one strand of the prespacer (fragment of foreign DNA) is integrated at the leader-repeat (R-L) junction of the CRISPR array. In the second step, the other strand of the prespacer integrates at the repeat-spacer (R-S) junction resulting in a fully integrated prespacer. The authors propose that, in vitro, water-mediated …
Reviewer #1 (Public Review):
In this manuscript Ma and colleagues present a biochemical investigation into integration by the Spy Cas1-Cas2 integrase, a key component of the type II CRISPR immune response. From this work they challenge the view that disintegration (the reverse of integration) only functions as an off-pathway reaction (proof reading) and propose that disintegration-promoted integration is functionally important for integration into CRISPR arrays.
CRISPR integration proceeds through two steps. In the first, one strand of the prespacer (fragment of foreign DNA) is integrated at the leader-repeat (R-L) junction of the CRISPR array. In the second step, the other strand of the prespacer integrates at the repeat-spacer (R-S) junction resulting in a fully integrated prespacer. The authors propose that, in vitro, water-mediated disintegration at the first site (R-L) is concomitant with second site integration (R-S). The authors present good evidence that this is happening in vitro though a series of elegant biochemical experiments using plasmid and oligo substrates.
This is an intriguing hypothesis but its significance is unclear. As the authors themselves acknowledge it could very well be that disintegration is not required in vivo and that this is an in vitro feature of the reaction. Admittedly, demonstrating that disintegration is important in vivo would be a very difficult task. In addition, the in vitro system used here is only partially reconstituted. The substrates lack a PAM sequence, which is necessary for protospacers to be incorporated in the correct orientation and may help direct the first integration event to the L-R junction. Presumably because of this all the reactions presented do not analyze the orientation of the incorporated prespacer sequence. Cas9 and Csn2 are also absent (as are other potentially required host factors), which are necessary for correct integration in vivo. Any of these missing components could change the behavior of the system with respect to disintegration.
-

Reviewer #2 (Public Review):
Ma and colleagues have show that when DNA fragments are inserted into a CRISPR array in vitro, single end integrations are the primary products of proto-spacer insertion and double-end insertions are rare. The reason for this outcome is that the second insertion event is accompanied by 3'-OH mediated disintegration or hydrolysis at the initial insertion site. A double-strand break would remain following filling of the repeat sequence gaps in vivo in the case of the hydrolysis products. The authors propose that hydrolysis of one end of the transposon DNA may be the primary mechanism for insertion of very small DNA elements (which will be difficult to bend tightly) as found for the proto spacer sequences and that cellular repair pathways would be responsible for ligating the CRISPR array back together.
The …
Reviewer #2 (Public Review):
Ma and colleagues have show that when DNA fragments are inserted into a CRISPR array in vitro, single end integrations are the primary products of proto-spacer insertion and double-end insertions are rare. The reason for this outcome is that the second insertion event is accompanied by 3'-OH mediated disintegration or hydrolysis at the initial insertion site. A double-strand break would remain following filling of the repeat sequence gaps in vivo in the case of the hydrolysis products. The authors propose that hydrolysis of one end of the transposon DNA may be the primary mechanism for insertion of very small DNA elements (which will be difficult to bend tightly) as found for the proto spacer sequences and that cellular repair pathways would be responsible for ligating the CRISPR array back together.
The manuscript is well written and the experiments carefully planned. The findings will be of interest to researchers studying transposition-like mechanisms in a broad range of systems. The experiments focus on analyses of Cas1-Cas2 catalyzed in vitro reactions using cleverly designed DNA substrates, leading to a hypothesis about how the system may work in vivo. The direct conclusions are well supported by the experimental data. The big unanswered question is whether the results obtained with a minimal Cas1-Cas2 system in vitro reflect what happens in the bacterial cell. More proteins are present in the cell that could modify the reaction and the native DNA array structure differs somewhat from what was used here. Overall, the results presented are intriguing, but their relationship to the cellular mechanism of CRISPR array generation is unclear, since it is not yet known whether DNA elements are inserted in vivo in the same way as observed here in vitro.
-

-