Faroese Whole Genomes Provide Insight into Ancestry and Recent Selection
Curation statements for this article:-
Curated by eLife
eLife Assessment
This paper presents an analysis of demography and selection from whole-genome sequencing of 40 Faroese, with data that are useful beyond the study region. Much of the analysis is solid, but a more fine-scale analysis of demographic history could have led to more interesting findings. In addition, there are concerns about the selection analyses, given the special nature of the studied population and sampling scheme. Finally, lack of data availability limits the broader value of the paper.
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Abstract
The Faroe Islands are home to descendants of a North Atlantic founder population with a unique history shaped by both migration and periods of relative isolation. Here, we investigate the genetic diversity, population structure, and demographic history of the islands by analyzing whole genome sequencing data from 40 participants in the Faroe Genome Project. This represents the first whole genome sequencing panel of this size from the Faroe Islands. We observed numerous putatively functional private alleles, including stop gain variants and high impact missense variants in the cohort. Faroese individuals had a higher proportion of their genomes contained in long runs of homozygosity than other European groups, including Finnish, suggesting a more recent or stronger bottleneck in the Faroese population. Signals of positive selection were identified at loci containing genes that play roles in vitamin D and dietary fat absorption and DNA repair, while increased diversity on lactase persistence haplotypes was observed. Fine-scale analysis of haplotype structure in modern and ancient European genomes revealed genetic affinities with ancient Iron Age individuals from the North and West of Europe, providing evidence for potential contributions to the Faroese gene pool from Celtic and Viking populations as well as information about the temporal order in which these events happened. This study highlights the impact of evolutionary processes, such as ancient admixture, founder events, and positive selection, on the present-day genetic architecture of North Atlantic founder populations like the Faroe Islands.
Article activity feed
-

Author response:
We thank the reviewers for their thoughtful comments and constructive suggestions. We describe how we will address each point below and are grateful for the guidance on areas where our work could be clarified or expanded. In particular, we note the following:
Selection scan summary statistics: In our revised manuscript, we will include summary statistics from the selection scans. We believe this addition will enhance transparency and provide additional context for readers.
Reporting of outliers: As highlighted by the editor, the reviewers expressed differing views on the most appropriate way to report outliers. To provide a comprehensive and balanced presentation, we will report both the empirical selection statistics and the corresponding converted p-values. This dual approach will allow readers to fully interpret the …
Author response:
We thank the reviewers for their thoughtful comments and constructive suggestions. We describe how we will address each point below and are grateful for the guidance on areas where our work could be clarified or expanded. In particular, we note the following:
Selection scan summary statistics: In our revised manuscript, we will include summary statistics from the selection scans. We believe this addition will enhance transparency and provide additional context for readers.
Reporting of outliers: As highlighted by the editor, the reviewers expressed differing views on the most appropriate way to report outliers. To provide a comprehensive and balanced presentation, we will report both the empirical selection statistics and the corresponding converted p-values. This dual approach will allow readers to fully interpret the results under both perspectives.
Methodological considerations: We have carefully considered the reviewers' methodological suggestions and will incorporate them into our revisions where possible. These changes strengthen the rigor and clarity of the analyses.
Public Reviews:
Reviewer #1 (Public review):
Summary:
The paper reports an analysis of whole-genome sequence data from 40 Faroese. The authors investigate aspects of demographic history and natural selection in this population. The key findings are that the Faroese (as expected) have a small population size and are broadly of Northwest European ancestry. Accordingly, selection signatures are largely shared with other Northwest European populations, although the authors identify signals that may be specific to the Faroes. Finally, they identify a few predicted deleterious coding variants that may be enriched in the Faroes.
Strengths:
The data are appropriately quality-controlled and appear to be of high quality. Some aspects of the Faroese population history are characterized, in particular, by the relatively (compared to other European populations) high proportion of long runs of homozygosity, which may be relevant for disease mapping of recessive variants. The selection analysis is presented reasonably, although as the authors point out, many aspects, for example differences in iHS, can reflect differences in demographic history or population-specific drift and thus can't reliably be interpreted in terms of differences in the strength of selection.
Weaknesses:
The main limitations of the paper are as follows:
(1) The data are not available. I appreciate that (even de-identified) genotype data cannot be shared; however, that does substantially reduce the value of the paper. Minimally, I think the authors should share summary statistics for the selection scans, in line with the standard of the field.
We agree with the reviewer that sharing the selection scan results is important, so in the next revision of this manuscript we will make the selection scan summary statistics publicly available, and clearly lay out the guidelines and research questions for which the data can be accessed.
(2) The insight into the population history of the Faroes is limited, relative to what is already known (i.e., they were settled around 1200 years ago, by people with a mixture of Scandinavian and British ancestry, have a small effective population size, and any admixture since then comes from substantially similar populations). It's obvious, for example, that the Faroese population has a smaller bottleneck than, say, GBR.
More sophisticated analyses (for example, ARG-based methods, or IBD or rare variant sharing) would be able to reveal more detailed and fine-scale information about the history of the populations that is not already known. PCA, ADMIXTURE, and HaplotNet analysis are broad summaries, but the interesting questions here would be more specific to the Faroes, for example, what are the proportions of Scandinavian vs Celtic ancestry? What is the date and extent of sex bias (as suggested by the uniparental data) in this admixture? I think that it is a bit of a missed opportunity not to address these questions.
We clarify that we did quantify the proportions of various ancestry components as estimated by HaploNet in main text Figure 5 and supplemental figures S5 and S6. In our revisions, we will include the average global ancestry of the various components in the Main Text so that this result is more clear.
We agree that more fine-scale demographic analyses would be informative. We have begun working on an estimation of the admixture date, for example, but have encountered problems with using different standard date estimation software, which give very inconsistent and unstable results. We suspect this might be due to the strong bottleneck experienced in the history of the Faroe Islands breaking one or more of the assumptions of these methods. We will continue working on this problem in coming months, possibly using simulations to assess where the problem might be. We recognize that our relatively small sample size places limits on the fine-scale demographic analyses that can be performed. We are addressing this in ongoing work by generating a larger cohort, which we hope will enable more detailed inference in the future.
(3) I don't really understand the rationale for looking at HLA-B allele frequencies. The authors write that "ankylosing spondylitis (AS) may be at a higher prevalence in the Faroe Islands (unpublished data), however, this has not been confirmed by follow-up epidemiological studies". So there's no evidence (certainly no published evidence) that AS is more prevalent, and hence nothing to explain with the HLA allele frequencies?
We agree that no published studies have confirmed a higher prevalence of ankylosing spondylitis (AS) in the Faroe Islands. Our recruitment data suggest that AS might be more common than in other European populations, but we understand that this is only based on limited, unpublished observations and what we are hearing from the community. We emphasized in our original manuscript that this is based on observational evidence from the FarGen project. However, as this reviewer pointed out, we can be more clear that this prevalence has not been formally studied.
In our next revision we will clarify in the text that our recruitment data suggest a higher prevalence of AS may be possible, but more formal epidemiological studies are needed to confirm this observation. The reason we study HLA-B allele frequencies is to see if the genetic background of the Faroese population could help explain this possible difference, since HLA-B27 is already known to play a strong role in AS.
Reviewer #2 (Public review):
In this paper, Hamid et al present 40 genomes from the Faroe Islands. They use these data (a pilot study for an anticipated larger-scale sequencing effort) to discuss the population genetic diversity and history of the sample, and the Faroes population. I think this is an overall solid paper; it is overall well-polished and well-written. It is somewhat descriptive (as might be expected for an explorative pilot study), but does make good use of the data.
The data processing and annotation follows a state-of-the-art protocol, and at least I could not find any evidence in the results that would pinpoint towards bioinformatic issues having substantially biased some of the results, and at least preliminary results lead to the identification of some candidate disease alleles, showing that small, isolated cohorts can be an efficient way to find populations with locally common, but globally rare disease alleles.
I also enjoyed the population structure analysis in the context of ancient samples, which gives some context to the genetic ancestry of Faroese, although it would have been nice if that could have been quantified, and it is unfortunate that the sampling scheme effectively precludes within-Faroes analyses.
We note that although the ancestry proportions are not specified in the main text, we did quantify ancestry proportions in the modern Faroese individuals and other ancient samples, and we visualized these proportions in Figure 5 and Supplementary Figures S5 and S6. As stated in our response to Reviewer #1, in our revisions, we will more clearly state the average global ancestry of the various components in the Main Text.
I am unfortunately quite critical of the selection analysis, both on a statistical level and, more importantly, I do not believe it measures what the authors think it does.
Major comments:
(1) Admixture timing/genomic scaling/localization:
As the authors lay out, the Faroes were likely colonized in the last 1,000-1,500 years, i.e., 40-60 generations ago. That means most genomic processes that have happened on the Faroese should have signatures that are on the order of ~1-2cM, whereas more local patterns likely indicate genetic history predating the colonization of the islands. Yet, the paper seems to be oblivious to this (to me) fascinating and somewhat unique premise. Maybe this thought is wrong, but I think the authors miss a chance here to explain why the reader should care beyond the fact that the small populations might have high-frequency risk alleles and the Faroes are intrinsically interesting, but more importantly, it also makes me think it leads to some misinterpretations in the selection analysis
See response to point #3
(2) ROH:
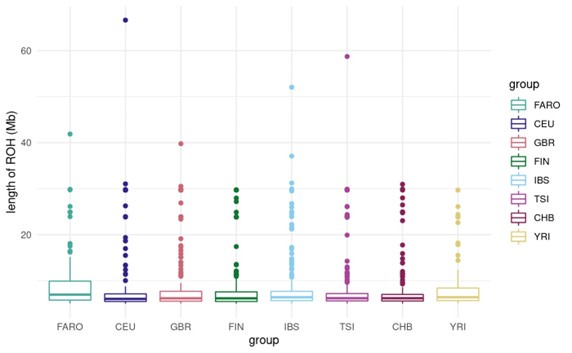
Would the sampling scheme impact ROH? How would it deal with individuals with known parental coancestry? As an example of what I mean by my previous comment, 1MB is short enough in that I would expect most/many 1MB ROH-tracts to come from pedigree loops predating the colonization of the Faroes. (i.e, I am actually quite surprised that there isn't much more long ROH, which makes me wonder if that would be impacted by the sampling scheme).
The sampling scheme was designed to choose 40 Faroese individuals that were representative of the different regions and were minimally related. There were no pairs of third-degree relatives or closer (pi-hat > 0.125) in either the Faroese cohort or the reference populations. It is possible that this sampling scheme would reduce the amount of longer ROHs in the population, but we should still be able to see overall patterns of ROH reflective of bottlenecks in the past tens of generations. Additionally, based on this reviewer's earlier comment, 1 Mb ROHs would still be relevant to demographic events in the last 40-60 generations given that on average 1 cM corresponds to 1 Mb in humans, though we recognize that is not an exact conversion.
That said, the “sum total amount of the genome contained in long ROH” as we described in the manuscript includes all ROHs greater than 1Mb. Although we group all ROHs longer than 1Mb into one category in the current manuscript, we can look more specifically at the distribution of the longer ROH in future revisions and add discussion into what this might tell us about the timing of bottlenecks.
For now, we share a plot of the distribution in ROH lengths across all individuals for each cohort. As this plot shows, there certainly are ROHs longer than 1Mb in the Faroese cohort, and on average there is a higher proportion of long ROH particularly in the 5-15 Mb range in the Faroese cohort relative to the other cohorts.
Author response image 1.
(3) Selection scan:
We are talking about a bottlenecked population that is recently admixed (Faroese), compared to a population (GBR) putatively more closely related to one of its sources. My guess would be that selection in such a scenario would be possibly very hard to detect, and even then, selection signals might not differentiate selection in Faroese vs. GBR, but rather selection/allele frequency differences between different source populations. I think it would be good to spell out why XP-EHH/iHS measures selection at the correct time scale, and how/if these statistics are expected to behave differently in an admixed population.
The reviewer brings up good points about the utility of classical selection statistics in populations that are admixed or bottlenecked, and whether the timescale at which these statistics detect selection is relevant for understanding the selective history of the Faroese population. We break down these concerns separately.
(1) Bottlenecks: Recent bottlenecks result in higher LD within a population. However, demographic events such as bottlenecks affect global genomic patterns while positive selection is expected to affect local genomic patterns. For this reason, iHS and XP-EHH statistics are standardized against the genome-wide background, to account for population-specific demographic history.
(2) Admixture: The term “admixture” has different interpretations depending on the line of inquiry and the populations being studied. Across various time and geographic scales, all human populations are admixed to some degree, as gene flow between groups is a common fixture throughout our history. For example,
even the modern British population has “admixed” ancestry from North / West European sources as well, dating to at least as recently as the Medieval & Viking periods (Gretzinger et al. 2022, Leslie et al. 2015), yet we do not commonly consider it an “admixed” population, and we are not typically concerned about applying haplotype-based statistics in this population. This is due to the low divergence between the source populations. In the case of the Faroe Islands, we believe admixture likely occurred on a similar timescale. We see low variance in ancestry proportions estimated by HaploNet, both from the historical Faroese individuals (250BP) and the modern samples. This indicates admixture predating the settlement of the Faroe Islands, where recombination has had time to break up long ancestry tracts and the global ancestry proportions have reached an equilibrium. That is, these ancestry patterns suggest that the modern Faroese are most likely descended from already admixed founders. We mention this as a likely possibility in the main text: “This could have occurred either via a mixture of the original “West Europe” ancestry with individuals of predominantly “North Europe” ancestry, or a by replacement with individuals that were already of mixed ancestry at the time of arrival in the islands (the latter are not uncommon in Viking Age mainland Europe).” And, as with the case of the British population, the closely-related ancestral sources for the Faroese founders were likely not so diverged as to have differences in allele frequencies and long-range haplotypes that would disrupt signals of selection from iHS or XP-EHH.
(3) Time scale: It is certainly possible, and in fact likely, that iHS measures selection older than the settlement of the Faroe Islands. In our manuscript, we calculated iHS in both the Faroese and the closely related British cohort, and we highlight in the main Main Text that the top signals, with the exception of LCT, are shared between the two cohorts, indicative of selection that began prior to the population split. iHS is a commonly calculated statistic, and it is often calculated in a single population without comparing to others, so we feel it is important to show our result demonstrating these shared selection signals. In future revisions, we will emphasize in the main text that we are not claiming to have identified selection that occurred in the Faroese population post-settlement with the iHS statistic. As far as XP-EHH, it is a statistic designed to identify differentiated variants that are fixed or approaching fixation in one population but not others. The time-scale of selection that XP-EHH can detect would therefore be dependent on the populations used for comparison. As XP-EHH has the best power to identify alleles that are fixed or approaching fixation in one population but not others, it is less likely to detect older selection events / incomplete sweeps from the source populations.
In our next revision, we will more clearly state limitations of these statistics under various population histories, and clarify the time-scale at which we are detecting selection for iHS vs XP-EHH.
(4) Similarly, for the discussion of LCT, I am not convinced that the haplotypes depicted here are on the right scale to reflect processes happening on the Faroes. Given the admixture/population history, it at the very least should be discussed in the context of whether the 13910 allele frequency on the Faroes is at odds with what would be expected based on the admixture sources.
We agree that more investigation into the LCT allele frequency in the other ancient samples may provide some insight into the selection history, particularly in light of ancient admixture. Please note, we did look at the allele frequency of the LCT allele rs4988235 and stated in the main text that it was present at high frequencies in the historical (250BP) Faroese samples. The frequency of this allele in the imputed historical Faroese samples is 82% while the allele is present at ~74% frequency in modern samples. We did not report the exact percentage in the main text because the sample size of the historical samples (11 individuals) is small and coverage of ancient samples is low, leading to potential errors in imputation. However, we can try to calculate the LCT allele frequency in other ancient samples, and assuming that we have good proxies for the sources at the time of admixture, we may calculate the expected allele frequency in the admixed ancestors of the Faroese founders in the next revision.
(5) I am lacking information to evaluate the procedure for turning the outliers into p-values. Both iHS and XP-EHH are ratio statistics, meaning they might be heavy-tailed if one is not careful, and the central limit theorem may not apply. It would be much easier (and probably sufficient for the points being made here) to reframe this analysis in terms of empirical outliers.
Given that there are disagreements on the best approach to reporting selection scan results from the reviewers, in our revision, we can additionally supply both the standardized iHS / XP-EHH values in the supplementary information as well as these values transformed to p-values. As the p-values are derived from the empirical distribution, the “significant” p-values are also empirical outliers from the empirical distribution, so the conclusions of the manuscript do not change. We found that the p-value approach and controlling for FDR is more conservative, with fewer signals reaching “significance” than are considered empirical outliers based on common approaches such as IQR or arbitrary percentile cutoffs.
(6) Oldest individual predating gene flow: It seems impossible to make any statements based on a single individual. Why is it implausible that this person (or their parents), e.g., moved to the Faroes within their lifetime and died there?
We agree with the reviewer that this is a plausible explanation, and in future revisions we will update the main text to acknowledge this possibility.
-

-

-

eLife Assessment
This paper presents an analysis of demography and selection from whole-genome sequencing of 40 Faroese, with data that are useful beyond the study region. Much of the analysis is solid, but a more fine-scale analysis of demographic history could have led to more interesting findings. In addition, there are concerns about the selection analyses, given the special nature of the studied population and sampling scheme. Finally, lack of data availability limits the broader value of the paper.
-

Reviewer #1 (Public review):
Summary:
The paper reports an analysis of whole-genome sequence data from 40 Faroese. The authors investigate aspects of demographic history and natural selection in this population. The key findings are that the Faroese (as expected) have a small population size and are broadly of Northwest European ancestry. Accordingly, selection signatures are largely shared with other Northwest European populations, although the authors identify signals that may be specific to the Faroes. Finally, they identify a few predicted deleterious coding variants that may be enriched in the Faroes.
Strengths:
The data are appropriately quality-controlled and appear to be of high quality. Some aspects of the Faroese population history are characterized, in particular, by the relatively (compared to other European populations) …
Reviewer #1 (Public review):
Summary:
The paper reports an analysis of whole-genome sequence data from 40 Faroese. The authors investigate aspects of demographic history and natural selection in this population. The key findings are that the Faroese (as expected) have a small population size and are broadly of Northwest European ancestry. Accordingly, selection signatures are largely shared with other Northwest European populations, although the authors identify signals that may be specific to the Faroes. Finally, they identify a few predicted deleterious coding variants that may be enriched in the Faroes.
Strengths:
The data are appropriately quality-controlled and appear to be of high quality. Some aspects of the Faroese population history are characterized, in particular, by the relatively (compared to other European populations) high proportion of long runs of homozygosity, which may be relevant for disease mapping of recessive variants. The selection analysis is presented reasonably, although as the authors point out, many aspects, for example differences in iHS, can reflect differences in demographic history or population-specific drift and thus can't reliably be interpreted in terms of differences in the strength of selection.
Weaknesses:
The main limitations of the paper are as follows:
(1) The data are not available. I appreciate that (even de-identified) genotype data cannot be shared; however, that does substantially reduce the value of the paper. Minimally, I think the authors should share summary statistics for the selection scans, in line with the standard of the field.
(2) The insight into the population history of the Faroes is limited, relative to what is already known (i.e., they were settled around 1200 years ago, by people with a mixture of Scandinavian and British ancestry, have a small effective population size, and any admixture since then comes from substantially similar populations). It's obvious, for example, that the Faroese population has a smaller bottleneck than, say, GBR.
More sophisticated analyses (for example, ARG-based methods, or IBD or rare variant sharing) would be able to reveal more detailed and fine-scale information about the history of the populations that is not already known. PCA, ADMIXTURE, and HaplotNet analysis are broad summaries, but the interesting questions here would be more specific to the Faroes, for example, what are the proportions of Scandinavian vs Celtic ancestry? What is the date and extent of sex bias (as suggested by the uniparental data) in this admixture? I think that it is a bit of a missed opportunity not to address these questions.
(3) I don't really understand the rationale for looking at HLA-B allele frequencies. The authors write that "ankylosing spondylitis (AS) may be at a higher prevalence in the Faroe Islands (unpublished data), however, this has not been confirmed by follow-up epidemiological studies". So there's no evidence (certainly no published evidence) that AS is more prevalent, and hence nothing to explain with the HLA allele frequencies?
-

Reviewer #2 (Public review):
In this paper, Hamid et al present 40 genomes from the Faroe Islands. They use these data (a pilot study for an anticipated larger-scale sequencing effort) to discuss the population genetic diversity and history of the sample, and the Faroes population. I think this is an overall solid paper; it is overall well-polished and well-written. It is somewhat descriptive (as might be expected for an explorative pilot study), but does make good use of the data.
The data processing and annotation follows a state-of-the-art protocol, and at least I could not find any evidence in the results that would pinpoint towards bioinformatic issues having substantially biased some of the results, and at least preliminary results lead to the identification of some candidate disease alleles, showing that small, isolated cohorts …
Reviewer #2 (Public review):
In this paper, Hamid et al present 40 genomes from the Faroe Islands. They use these data (a pilot study for an anticipated larger-scale sequencing effort) to discuss the population genetic diversity and history of the sample, and the Faroes population. I think this is an overall solid paper; it is overall well-polished and well-written. It is somewhat descriptive (as might be expected for an explorative pilot study), but does make good use of the data.
The data processing and annotation follows a state-of-the-art protocol, and at least I could not find any evidence in the results that would pinpoint towards bioinformatic issues having substantially biased some of the results, and at least preliminary results lead to the identification of some candidate disease alleles, showing that small, isolated cohorts can be an efficient way to find populations with locally common, but globally rare disease alleles.
I also enjoyed the population structure analysis in the context of ancient samples, which gives some context to the genetic ancestry of Faroese, although it would have been nice if that could have been quantified, and it is unfortunate that the sampling scheme effectively precludes within-Faroes analyses.
I am unfortunately quite critical of the selection analysis, both on a statistical level and, more importantly, I do not believe it measures what the authors think it does.
Major comments:
(1) Admixture timing/genomic scaling/localization:
As the authors lay out, the Faroes were likely colonized in the last 1,000-1,500 years, i.e., 40-60 generations ago. That means most genomic processes that have happened on the Faroese should have signatures that are on the order of ~1-2cM, whereas more local patterns likely indicate genetic history predating the colonization of the islands. Yet, the paper seems to be oblivious to this (to me) fascinating and somewhat unique premise. Maybe this thought is wrong, but I think the authors miss a chance here to explain why the reader should care beyond the fact that the small populations might have high-frequency risk alleles and the Faroes are intrinsically interesting, but more importantly, it also makes me think it leads to some misinterpretations in the selection analysis(2) ROH:
Would the sampling scheme impact ROH? How would it deal with individuals with known parental coancestry? As an example of what I mean by my previous comment, 1MB is short enough in that I would expect most/many 1MB ROH-tracts to come from pedigree loops predating the colonization of the Faroes. (i.e, I am actually quite surprised that there isn't much more long ROH, which makes me wonder if that would be impacted by the sampling scheme).(3) Selection scan:
We are talking about a bottlenecked population that is recently admixed (Faroese), compared to a population (GBR) putatively more closely related to one of its sources. My guess would be that selection in such a scenario would be possibly very hard to detect, and even then, selection signals might not differentiate selection in Faroese vs. GBR, but rather selection/allele frequency differences between different source populations. I think it would be good to spell out why XP-EHH/iHS measures selection at the correct time scale, and how/if these statistics are expected to behave differently in an admixed population.
(4) Similarly, for the discussion of LCT, I am not convinced that the haplotypes depicted here are on the right scale to reflect processes happening on the Faroes. Given the admixture/population history, it at the very least should be discussed in the context of whether the 13910 allele frequency on the Faroes is at odds with what would be expected based on the admixture sources.
(5) I am lacking information to evaluate the procedure for turning the outliers into p-values. Both iHS and XP-EHH are ratio statistics, meaning they might be heavy-tailed if one is not careful, and the central limit theorem may not apply. It would be much easier (and probably sufficient for the points being made here) to reframe this analysis in terms of empirical outliers.
(6) Oldest individual predating gene flow: It seems impossible to make any statements based on a single individual. Why is it implausible that this person (or their parents), e.g., moved to the Faroes within their lifetime and died there?
-