Functionally coupled ion channels begin co-assembling at the start of their synthesis
Curation statements for this article:-
Curated by eLife
eLife Assessment
This fundamental manuscript provides compelling evidence that BK and CaV1.3 channels can co-localize as ensembles early in the biosynthetic pathway, including within the ER and Golgi. The findings, supported by a range of imaging and proximity assays, offer insights into channel organization in both heterologous and endogenous systems. The data substantiate the central claims, while highlighting intriguing mechanistic questions for future studies: the determinants of mRNA co-localization, the temporal dynamics of ensemble trafficking, and the physiological implications of pre-assembly for channel function at the plasma membrane.
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Reading List (BiophysicsColab)
- Evaluated articles (eLife)
Abstract
Calcium binding to BK channels lowers BK activation threshold, substantiating functional coupling with calcium-permeable channels. This coupling requires close proximity between different channel types, and the formation of BK-Ca V 1.3 hetero-clusters at nanometer distances exemplifies this unique organization. To investigate the structural basis of this interaction, we tested the hypothesis that BK and Ca V 1.3 channels assemble before their insertion into the plasma membrane. Our approach incorporated four strategies: (1) detecting interactions between BK and Ca V 1.3 proteins inside the cell, (2) identifying membrane compartments where intracellular hetero-clusters reside, (3) measuring the proximity of their mRNAs, and (4) assessing protein interactions at the plasma membrane during early translation. These analyses revealed that a subset of BK and Ca V 1.3 transcripts are spatially close in micro-translational complexes, and their newly synthesized proteins associate within the endoplasmic reticulum (ER) and Golgi. Comparisons with other proteins, transcripts, and randomized localization models support the conclusion that BK and Ca V 1.3 hetero-clusters form before their insertion at the plasma membrane.
Article activity feed
-

-

-

-

eLife Assessment
This fundamental manuscript provides compelling evidence that BK and CaV1.3 channels can co-localize as ensembles early in the biosynthetic pathway, including within the ER and Golgi. The findings, supported by a range of imaging and proximity assays, offer insights into channel organization in both heterologous and endogenous systems. The data substantiate the central claims, while highlighting intriguing mechanistic questions for future studies: the determinants of mRNA co-localization, the temporal dynamics of ensemble trafficking, and the physiological implications of pre-assembly for channel function at the plasma membrane.
-

Reviewer #1 (Public review):
Summary:
The co-localization of large conductance calcium- and voltage activated potassium (BK) channels with voltage-gated calcium channels (CaV) at the plasma membrane is important for the functional role of these channels in controlling cell excitability and physiology in a variety of systems. An important question in the field is where and how do BK and CaV channels assemble as 'ensembles' to allow this coordinated regulation - is this through preassembly early in the biosynthetic pathway, during trafficking to the cell surface or once channels are integrated into the plasma membrane. These questions also have broader implications for assembly of other ion channel complexes. Using an imaging based approach, this paper addresses the spatial distribution of BK-CaV ensembles using both overexpression …
Reviewer #1 (Public review):
Summary:
The co-localization of large conductance calcium- and voltage activated potassium (BK) channels with voltage-gated calcium channels (CaV) at the plasma membrane is important for the functional role of these channels in controlling cell excitability and physiology in a variety of systems. An important question in the field is where and how do BK and CaV channels assemble as 'ensembles' to allow this coordinated regulation - is this through preassembly early in the biosynthetic pathway, during trafficking to the cell surface or once channels are integrated into the plasma membrane. These questions also have broader implications for assembly of other ion channel complexes. Using an imaging based approach, this paper addresses the spatial distribution of BK-CaV ensembles using both overexpression strategies in tsa201 and INS-1 cells and analysis of endogenous channels in INS-1 cells using proximity ligation and superesolution approaches. In addition, the authors analyse the spatial distribution of mRNAs encoding BK and Cav1.3. The key conclusion of the paper that BK and CaV1.3 are co-localised as ensembles intracellularly in the ER and Golgi is well supported by the evidence. The experiments and analysis are carefully performed and the findings are very well presented.
-

Reviewer #3 (Public review):
Summary:
The authors present a clearly written and beautifully presented piece of work demonstrating clear evidence to support the idea that BK channels and Cav1.3 channels can co-assemble prior to their assertion in the plasma membrane.
Strengths:
The experimental records shown back up their hypotheses and the authors are to be congratulated for the large number of control experiments shown in the ms.
-

Author response:
The following is the authors’ response to the previous reviews.
Reviewer #1 (Public review):
Summary:
This manuscript by Pournejati et al investigates how BK (big potassium) channels and CaV1.3 (a subtype of voltage-gated calcium channels) become functionally coupled by exploring whether their ensembles form early-during synthesis and intracellular trafficking-rather than only after insertion into the plasma membrane. To this end, the authors use the PLA technique to assess the formation of ion channel associations in the different compartments (ER, Golgi or PM), single-molecule RNA in situ hybridization (RNAscope), and super-resolution microscopy.
Strengths:
The manuscript is well written and addresses an interesting question, combining a range of imaging techniques. The findings are generally well-presented and offer …
Author response:
The following is the authors’ response to the previous reviews.
Reviewer #1 (Public review):
Summary:
This manuscript by Pournejati et al investigates how BK (big potassium) channels and CaV1.3 (a subtype of voltage-gated calcium channels) become functionally coupled by exploring whether their ensembles form early-during synthesis and intracellular trafficking-rather than only after insertion into the plasma membrane. To this end, the authors use the PLA technique to assess the formation of ion channel associations in the different compartments (ER, Golgi or PM), single-molecule RNA in situ hybridization (RNAscope), and super-resolution microscopy.
Strengths:
The manuscript is well written and addresses an interesting question, combining a range of imaging techniques. The findings are generally well-presented and offer important insights into the spatial organization of ion channel complexes, both in heterologous and endogenous systems.
Weaknesses:
The authors have improved their manuscript after revisions, and some previous concerns have been addressed.
Still, the main concern about this work is that the current experiments do not quantitatively or mechanistically link the ensembles observed intracellularly (in the endoplasmic reticulum (ER) or Golgi) to those found at the plasma membrane (PM). As a result, it is difficult to fully integrate the findings into a coherent model of trafficking. Specifically, the manuscript does not address what proportion of ensembles detected at the PM originated in the ER. Without data on the turnover or halflife of these ensembles at the PM, it remains unclear how many persist through trafficking versus forming de novo at the membrane. The authors report the percentage of PLApositive ensembles localized to various compartments, but this only reflects the distribution of pre-formed ensembles. What remains unknown is the proportion of total BK and CaV1.3 channels (not just those in ensembles) that are engaged in these complexes within each compartment. Without this, it is difficult to determine whether ensembles form in the ER and are then trafficked to the PM, or if independent ensemble formation also occurs at the membrane. To support the model of intracellular assembly followed by coordinated trafficking, it would be important to quantify the fraction of the total channel population that exists as ensembles in each compartment. A comparable ensemble-to-total ratio across ER and PM would strengthen the argument for directed trafficking of pre-assembled channel complexes.
We appreciate the reviewer’s thoughtful comment and agree that quantitatively linking intracellular hetero-clusters to those at the plasma membrane is an important and unresolved question. Our current study does not determine what proportion of ensembles at the plasma membrane originated during trafficking. It also does not quantify the fraction of total BK and CaV1.3 channels engaged in these complexes within each compartment. Addressing this requires simultaneous measurement of multiple parameters—total BK channels, total CaV1.3 channels, hetero-cluster formation (via PLA), and compartment identity—in the same cell. This is technically challenging. The antibodies used for channel detection are also required for the proximity ligation assay, which makes these measurements incompatible within a single experiment.
To overcome these limitations, we are developing new genetically encoded tools to enable real-time tracking of BK and CaV1.3 dynamics in live cells. These approaches will enable us to monitor channel trafficking and the formation of hetero-clusters, as detected by colocalization. This kind of experiments will provide insight into their origin and turnover. While these experiments are beyond the scope of the current study, the findings in our current manuscript provide the first direct evidence that BK and CaV channels can form hetero-clusters intracellularly prior to reaching the plasma membrane. This mechanistic insight reveals a previously unrecognized step in channel organization and lays the foundation for future work aimed at quantifying ensemble-to-total ratios and determining whether coordinated trafficking of pre-assembled complexes occurs.
This limitation is acknowledged in the discussion section, page 23. It reads: “Our findings highlight the intracellular assembly of BK-CaV1.3 hetero-clusters, though limitations in resolution and organelle-specific analysis prevent precise quantification of the proportion of intracellular complexes that ultimately persist on the cell surface.”
Reviewer #2 (Public review):
Summary:
The co-localization of large conductance calcium- and voltage activated potassium (BK) channels with voltage-gated calcium channels (CaV) at the plasma membrane is important for the functional role of these channels in controlling cell excitability and physiology in a variety of systems.
An important question in the field is where and how do BK and CaV channels assemble as 'ensembles' to allow this coordinated regulation - is this through preassembly early in the biosynthetic pathway, during trafficking to the cell surface or once channels are integrated into the plasma membrane. These questions also have broader implications for assembly of other ion channel complexes
Using an imaging based approach, this paper addresses the spatial distribution of BKCaV ensembles using both overexpression strategies in tsa201 and INS-1 cells and analysis of endogenous channels in INS-1 cells using proximity ligation and superesolution approaches. In addition, the authors analyse the spatial distribution of mRNAs encoding BK and Cav1.3.
The key conclusion of the paper that BK and CaV1.3 are co-localised as ensembles intracellularly in the ER and Golgi is well supported by the evidence.However, whether they are preferentially co-translated at the ER, requires further work. Moreover, whether intracellular pre-assembly of BK-CaV1.3 complexes is the major mechanism for functional complexes at the plasma membrane in these models requires more definitive evidence including both refinement of analysis of current data as well as potentially additional experiments.
The reviewer raises the question of whether BK and CaV1.3 channels are preferentially co-translated. In fact, I would like to propose that co-translation has not yet been clearly defined for this type of interaction between ion channels. In our current work, we 1) observed the colocalization between BK and CaV1.3 mRNAs and 2) determined that 70% of BK mRNA in active translation also colocalizes with CaV1.3 mRNA. We think these results favor the idea of translational complexes that can underlie the process of co-translation. However, and in total agreement with the Reviewer, the conclusion that the mRNA for the two ion channels is cotranslated would require further experimentation. For instance, mRNA coregulation is one aspect that could help to define co-translation.
To avoid overinterpretation, we have revised the manuscript to remove references to “co-translation” in the Results section and included the word “potential” when referring to co-translation in the Discussion section. We also clarified the limitations of our evidence in the Discussion that can be found on page 25: “It is important to note that while our data suggest mRNA coordination, additional experiments are required to directly assess co-translation.”
Strengths & Weaknesses
(1) Using proximity ligation assays of overexpressed BK and CaV1.3 in tsa201 and INS1 cells the authors provide strong evidence that BK and CaV can exist as ensembles (ie channels within 40 nm) at both the plasma membrane and intracellular membranes, including ER and Golgi. They also provide evidence for endogenous ensemble assembly at the Golgi in INS-1 cells and it would have been useful to determine if endogenous complexes are also observe in the ER of INS-1 cells. There are some useful controls but the specificity of ensemble formation would be better determined using other transmembrane proteins rather than peripheral proteins (eg Golgi 58K).
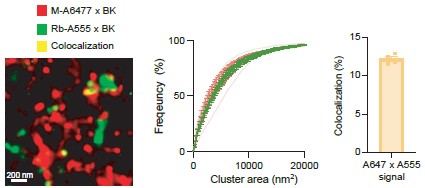
We thank the reviewer for their thoughtful feedback and for recognizing the strength of our proximity ligation assay data supporting BK–CaV1.3 hetero-clusters formation at both the plasma membrane and intracellular compartments. As for specificity controls, we appreciate the suggestion to use transmembrane markers. To strengthen our conclusion, we have performed an additional experiment comparing the number of PLA puncta formed by the interaction of CaV1.3 and BK channels with the number of PLA puncta formed by the interaction of CaV1.3 channels and ryanodine receptors in INS-1 cells. As shown in the figure below, the number of interactions between CaV1.3 and BK channels is significantly higher than that between CaV1.3 and RyR2. Of note, RyR2 is a protein resident of the ER. These results provide additional evidence of the existence of endogenous complex formation in INS-1 cells. We have added this figure as a supplement.
(2) Ensemble assembly was also analysed using super-resolution (dSTORM) imaging in INS-1 cells. In these cells only 7.5% of BK and CaV particles (endogenous?) co-localise that was only marginally above chance based on scrambled images. More detailed quantification and validation of potential 'ensembles' needs to be made for example by exploring nearest neighbour characteristics (but see point 4 below) to define proportion of ensembles versus clusters of BK or Cav1.3 channels alone etc. For example, it is mentioned that a distribution of distances between BK and Cav is seen but data are not shown.
We thank the reviewer for this comment. To address the request for more detailed quantification and validation of ensembles, we performed additional analyses:
Proportion of ensembles vs isolated clusters: We quantified clusters within 200 nm and found that 37 ± 3% of BK clusters are near one or more CaV1.3 clusters, whereas 15 ± 2% of CaV1.3 clusters are near BK clusters. Figure 8– Supplementary 1A
Distance distribution: As shown in Figure 8–Supplementary 1B, the nearestneighbor distance distribution for BK-to-CaV1.3 in INS-1 cells (magenta) is shifted toward shorter distances compared to randomized controls (gray), supporting preferential localization of BK–CaV1.3 hetero-clusters.
Together, these analyses confirm that BK–CaV1.3 ensembles occur more frequently than expected by chance and exhibit an asymmetric organization favoring BK proximity to CaV1.3 in INS-1 cells. We have included these data and figures in the revised manuscript, as well as description in the Results section.
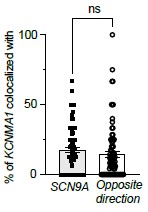
(3) The evidence that the intracellular ensemble formation is in large part driven by cotranslation, based on co-localisation of mRNAs using RNAscope, requires additional critical controls and analysis. The authors now include data of co-localised BK protein that is suggestive but does not show co-translation. Secondly, while they have improved the description of some controls mRNA co-localisation needs to be measured in both directions (eg BK - SCN9A as well as SCN9A to BK) especially if the mRNAs are expressed at very different levels. The relative expression levels need to be clearly defined in the paper. Authors also use a randomized image of BK mRNA to show specificity of co-localisation with Cav1.3 mRNA, however the mRNA distribution would not be expected to be random across the cell but constrained by ER morphology if cotranslated so using ER labelling as a mask would be useful?
We thank the reviewer for these constructive suggestions. We measured mRNA colocalization in both directions as recommended. As shown in the figure below, colocalization between KCNMA1 and SCN9A transcripts was comparable in both directions, with no statistically significant difference, supporting the specificity of the observed associations. We decided not to add this to the original figure to keep the figure simple.
We agree that co-localization of BK protein with BK mRNA is not conclusive evidence of co-translation, and we do not intend to mislead readers in our conclusion. Consequently, we were careful in avoiding the use of co-translation in the result section and added the word “potential” when referring to co-translation in the Discussion section. We added a sentence in the discussion to caution our interpretation: “It is important to note that while our data suggest mRNA coordination, additional experiments are required to directly assess cotranslation.”
Author response image 1.
(4) The authors attempt to define if plasma membrane assemblies of BK and CaV occur soon after synthesis. However, because the expression of BK and CaV occur at different times after transient transfection of plasmids more definitive experiments are required. For example, using inducible constructs to allow precise and synchronised timing of transcription. This would also provide critical evidence that co-assembly occurs very early in synthesis pathways - ie detecting complexes at ER before any complexes
We appreciate the reviewer’s insightful suggestion regarding the use of inducible constructs to synchronize transcription timing. This is an excellent approach and would allow direct testing of whether co-assembly occurs early in the synthesis pathway, including detection of complexes at the ER prior to plasma membrane localization. These experiments are beyond the scope of the present work but represent an important direction for future studies.
We have added the following sentence to the Discussion section (page 24) to highlight this idea. “Future experiments using inducible constructs to precisely control transcription timing will enable more precise quantification of heterocluster formation in the ER compartment prior to plasma membrane insertion and reduce the variability introduced by differences in expression timing after plasmid transfection.”
(5) While the authors have improved the definition of hetero-clusters etc it is still not clear in superesolution analysis, how they separate a BK tetramer from a cluster of BK tetramers with the monoclonal antibody employed ie each BK channel will have 4 binding sites (4 subunits in tetramer) whereas Cav1.3 has one binding site per channel. Thus, how do authors discriminate between a single BK tetramer (molecular cluster) with potential 4 antibodies bound compared to a cluster of 4 independent BK channels.
We appreciate the reviewer’s thoughtful comment regarding the interpretation of super-resolution data. We agree that distinguishing a single BK tetramer from a cluster of multiple BK channels is challenging when using an antibody that can bind up to four sites per channel. To clarify, our analysis does not attempt to resolve individual subunits within a tetramer; rather, it focuses on the nanoscale spatial proximity of BK and CaV1.3 signals.
We want to note that this limitation applies only to the super-resolution maps in Figures 8C and 9D and does not affect Airyscan-based analyses or measurements of BK–CaV1.3 proximity.
To address how we might distinguish between a single BK tetramer and a cluster of multiple BK channels, we considered two contrasting scenarios. In the first case, we assume that all four α-subunits within a tetramer are labeled. Based on cryoEM structures, a BK tetramer measures approximately 13 nm × 13 nm (≈169 nm²). Adding two antibody layers (primary and secondary) would increase the footprint by ~14 nm in each direction, resulting in an estimated area of ~41 nm × 41 nm (≈1681 nm²). Under this assumption, particles smaller than ~1681 nm² would likely represent individual tetramers, whereas larger particles would correspond to clusters of multiple tetramers.
In the second scenario, we propose that steric constraints at the S9–S10 segment, where the antibody binds, limit labeling to a single antibody per tetramer. If true, the localization precision would approximate 14 nm × 14 nm—the combined size of the antibody complex and the channel—close to the resolution limit of the microscope. To test this, we performed a control experiment using two antibodies targeting the BK C-terminal domain, raised in different species and labeled with distinct fluorophores. Super-resolution imaging revealed that only ~12% of particles were colocalized, suggesting that most channels bind a single antibody.
If multiple antibodies could bind each tetramer, we would expect much greater colocalization.
Although these data are not included in the manuscript, we have added the following clarification to the Results section (page 19): “It is important to note that this technique does not allow us to distinguish between labeling of four BK αsubunits within a tetramer and labeling of multiple BK channel clusters. Hence, particles smaller than ~1680 nm² may represent either a single tetramer or a cluster. This limitation applies to Figures 8C and 9D and does not affect measurements of BK–CaV1.3 proximity.”
Author response image 2.
(6) The post-hoc tests used for one way ANOVA and ANOVA statistics need to be defined throughout
We thank the reviewer for highlighting the need for clarity regarding our statistical analyses. We have now specified the post-hoc tests used for all one-way ANOVA and ANOVA comparisons throughout the manuscript, and updated figure legends.
Reviewer #3 (Public review):
Summary:
The authors present a clearly written and beautifully presented piece of work demonstrating clear evidence to support the idea that BK channels and Cav1.3 channels can co-assemble prior to their assertion in the plasma membrane.
Strengths:
The experimental records shown back up their hypotheses and the authors are to be congratulated for the large number of control experiments shown in the ms.
Recommendations for the authors:
Reviewer #1 (Recommendations for the authors):
The authors have sufficiently addressed the specific points previously raised and the manuscript has improved clarity in those aspects. My main concern, which still remains, is stated in the public review.
Reviewer #3 (Recommendations for the authors):
I am content that the authors have attempted to fully address my previous criticisms.
I have only three suggestions
(1) I think the word Homo-clusters at the bottom right of Figure 1 is erroneously included.
We thank the reviewer for bringing this to our attention. The figure has been corrected accordingly.
(2) The authors should, for completeness, to refer to the beta, gamma and LINGO subunit families in the Introduction and include appropriate references:
Knaus, H. G., Folander, K., Garcia-Calvo, M., Garcia, M. L., Kaczorowski, G. J., Smith, M., & Swanson, R. (1994). Primary sequence and immunological characterization of betasubunit of high conductance Ca2+-activated K+ channel from smooth muscle. The Journal of Biological Chemistry, 269(25), 17274-17278.
Brenner, R., Jegla, T. J., Wickenden, A., Liu, Y., & Aldrich, R. W. (2000a). Cloning and functional characterization of novel large conductance calcium-activated potassium channel beta subunits, hKCNMB3 and hKCNMB4. The Journal of Biological Chemistry, 275(9), 6453-6461.
Yan, J & R.W. Aldrich. (2010) LRRC26 auxiliary protein allows BK channel activation at resting voltage without calcium. Nature. 466(7305):513-516
Yan, J & R.W. Aldrich. (2012) BK potassium channel modulation by leucine-rich repeatcontaining proteins. Proceedings of the National Academy of Sciences 109(20):7917-22
Dudem, S, Large RJ, Kulkarni S, McClafferty H, Tikhonova IG, Sergeant, GP, Thornbury, KD, Shipston, MJ, Perrino BA & Hollywood MA (2020). LINGO1 is a novel regulatory subunit of large conductance, Ca2+-activated potassium channels. Proceedings of the National Academy of Sciences 117 (4) 2194-2200
Dudem, S., Boon, P. X., Mullins, N., McClafferty, H., Shipston, M. J., Wilkinson, R. D. A., Lobb, I., Sergeant, G. P., Thornbury, K. D., Tikhonova, I. G., & Hollywood, M. A. (2023). Oxidation modulates LINGO2-induced inactivation of large conductance, Ca2+-activated potassium channels. The Journal of Biological Chemistry, 299 (3) 102975.
We agree with the reviewer’s suggestion and have revised the Introduction to include references to the beta, gamma, and LINGO subunit families. Appropriate citations have been added to ensure completeness and contextual relevance.
Additionally, BK channels are modulated by auxiliary subunits, which fine-tune BK channel gating properties to adapt to different physiological conditions. The β, γ, and LINGO1 subunits each contribute distinct structural and regulatory features: β-subunits modulate Ca²⁺ sensitivity and can induce inactivation; γ-subunits shift voltage-dependent activation to more negative potentials; and LINGO1 reduces surface expression and promotes rapid inactivation (18-24). These interactions ensure precise control over channel activity, allowing BK channels to integrate voltage and calcium signals dynamically in various cell types.
(3) I think it may be more appropriate to include the sentence "The probes against the mRNAs of interest and tested in this work were designed by Advanced Cell Diagnostics." (P16, right hand column, L12-14) in the appropriate section of the Methods, rather than in Results.
We thank the reviewer for this helpful suggestion. In response, we have relocated the sentence to the appropriate section of the Methods, where it now appears with relevant context.
-

eLife Assessment
This important manuscript provides compelling evidence that BK and CaV1.3 channels can co-localize as ensembles early in the biosynthetic pathway, including in the ER and Golgi. The findings, supported by a range of imaging and proximity assays, offer insights into channel organization in both heterologous and endogenous systems. While the data broadly support the central claims, mechanistic aspects remain unresolved, particularly regarding the determinants of mRNA co-localization, the temporal dynamics of ensemble trafficking, and the physiological implications of pre-assembly for channel function at the plasma membrane.
-

Reviewer #1 (Public review):
Summary:
This manuscript by Pournejati et al investigates how BK (big potassium) channels and CaV1.3 (a subtype of voltage-gated calcium channels) become functionally coupled by exploring whether their ensembles form early-during synthesis and intracellular trafficking-rather than only after insertion into the plasma membrane. To this end, the authors use the PLA technique to assess the formation of ion channel associations in the different compartments (ER, Golgi or PM), single-molecule RNA in situ hybridization (RNAscope), and super-resolution microscopy.
Strengths:
The manuscript is well written and addresses an interesting question, combining a range of imaging techniques. The findings are generally well-presented and offer important insights into the spatial organization of ion channel complexes, both …
Reviewer #1 (Public review):
Summary:
This manuscript by Pournejati et al investigates how BK (big potassium) channels and CaV1.3 (a subtype of voltage-gated calcium channels) become functionally coupled by exploring whether their ensembles form early-during synthesis and intracellular trafficking-rather than only after insertion into the plasma membrane. To this end, the authors use the PLA technique to assess the formation of ion channel associations in the different compartments (ER, Golgi or PM), single-molecule RNA in situ hybridization (RNAscope), and super-resolution microscopy.
Strengths:
The manuscript is well written and addresses an interesting question, combining a range of imaging techniques. The findings are generally well-presented and offer important insights into the spatial organization of ion channel complexes, both in heterologous and endogenous systems.
Weaknesses:
The authors have improved their manuscript after revisions, and some previous concerns have been addressed. Still, the main concern about this work is that the current experiments do not quantitatively or mechanistically link the ensembles observed intracellularly (in the endoplasmic reticulum (ER) or Golgi) to those found at the plasma membrane (PM). As a result, it is difficult to fully integrate the findings into a coherent model of trafficking. Specifically, the manuscript does not address what proportion of ensembles detected at the PM originated in the ER. Without data on the turnover or half-life of these ensembles at the PM, it remains unclear how many persist through trafficking versus forming de novo at the membrane. The authors report the percentage of PLA-positive ensembles localized to various compartments, but this only reflects the distribution of pre-formed ensembles. What remains unknown is the proportion of total BK and CaV1.3 channels (not just those in ensembles) that are engaged in these complexes within each compartment. Without this, it is difficult to determine whether ensembles form in the ER and are then trafficked to the PM, or if independent ensemble formation also occurs at the membrane. To support the model of intracellular assembly followed by coordinated trafficking, it would be important to quantify the fraction of the total channel population that exists as ensembles in each compartment. A comparable ensemble-to-total ratio across ER and PM would strengthen the argument for directed trafficking of pre-assembled channel complexes.
-

Reviewer #2 (Public review):
Summary:
The co-localization of large conductance calcium- and voltage activated potassium (BK) channels with voltage-gated calcium channels (CaV) at the plasma membrane is important for the functional role of these channels in controlling cell excitability and physiology in a variety of systems.
An important question in the field is where and how do BK and CaV channels assemble as 'ensembles' to allow this coordinated regulation - is this through preassembly early in the biosynthetic pathway, during trafficking to the cell surface or once channels are integrated into the plasma membrane. These questions also have broader implications for assembly of other ion channel complexes.
Using an imaging based approach, this paper addresses the spatial distribution of BK-CaV ensembles using both overexpression …
Reviewer #2 (Public review):
Summary:
The co-localization of large conductance calcium- and voltage activated potassium (BK) channels with voltage-gated calcium channels (CaV) at the plasma membrane is important for the functional role of these channels in controlling cell excitability and physiology in a variety of systems.
An important question in the field is where and how do BK and CaV channels assemble as 'ensembles' to allow this coordinated regulation - is this through preassembly early in the biosynthetic pathway, during trafficking to the cell surface or once channels are integrated into the plasma membrane. These questions also have broader implications for assembly of other ion channel complexes.
Using an imaging based approach, this paper addresses the spatial distribution of BK-CaV ensembles using both overexpression strategies in tsa201 and INS-1 cells and analysis of endogenous channels in INS-1 cells using proximity ligation and superesolution approaches. In addition, the authors analyse the spatial distribution of mRNAs encoding BK and Cav1.3.
The key conclusion of the paper that BK and CaV1.3 are co-localised as ensembles intracellularly in the ER and Golgi is well supported by the evidence. However, whether they are preferentially co-translated at the ER, requires further work. Moreover, whether intracellular pre-assembly of BK-CaV complexes is the major mechanism for functional complexes at the plasma membrane in these models requires more definitive evidence including both refinement of analysis of current data as well as potentially additional experiments.
Strengths & Weaknesses
(1) Using proximity ligation assays of overexpressed BK and CaV1.3 in tsa201 and INS-1 cells the authors provide strong evidence that BK and CaV can exist as ensembles (ie channels within 40 nm) at both the plasma membrane and intracellular membranes, including ER and Golgi. They also provide evidence for endogenous ensemble assembly at the Golgi in INS-1 cells and it would have been useful to determine if endogenous complexes are also observe in the ER of INS-1 cells. There are some useful controls but the specificity of ensemble formation would be better determined using other transmembrane proteins rather than peripheral proteins (eg Golgi 58K).
(2) Ensemble assembly was also analysed using super-resolution (dSTORM) imaging in INS-1 cells. In these cells only 7.5% of BK and CaV particles (endogenous?) co-localise that was only marginally above chance based on scrambled images. More detailed quantification and validation of potential 'ensembles' needs to be made for example by exploring nearest neighbour characteristics (but see point 4 below) to define proportion of ensembles versus clusters of BK or Cav1.3 channels alone etc. For example, it is mentioned that a distribution of distances between BK and Cav is seen but data are not shown.
(3) The evidence that the intracellular ensemble formation is in large part driven by co-translation, based on co-localisation of mRNAs using RNAscope, requires additional critical controls and analysis. The authors now include data of co-localised BK protein that is suggestive but does not show co-translation. Secondly, while they have improved the description of some controls mRNA co-localisation needs to be measured in both directions (eg BK - SCN9A as well as SCN9A to BK) especially if the mRNAs are expressed at very different levels. The relative expression levels need to be clearly defined in the paper. Authors also use a randomized image of BK mRNA to show specificity of co-localisation with Cav1.3 mRNA, however the mRNA distribution would not be expected to be random across the cell but constrained by ER morphology if co-translated so using ER labelling as a mask would be useful?
(4) The authors attempt to define if plasma membrane assemblies of BK and CaV occur soon after synthesis. However, because the expression of BK and CaV occur at different times after transient transfection of plasmids more definitive experiments are required. For example, using inducible constructs to allow precise and synchronised timing of transcription. This would also provide critical evidence that co-assembly occurs very early in synthesis pathways - ie detecting complexes at ER before any complexes at Golgi or plasma membrane.
(5) While the authors have improved the definition of hetero-clusters etc it is still not clear in superesolution analysis, how they separate a BK tetramer from a cluster of BK tetramers with the monoclonal antibody employed ie each BK channel will have 4 binding sites (4 subunits in tetramer) whereas Cav1.3 has one binding site per channel. Thus, how do authors discriminate between a single BK tetramer (molecular cluster) with potential 4 antibodies bound compared to a cluster of 4 independent BK channels.
(6) The post-hoc tests used for one way ANOVA and ANOVA statistics need to be defined throughout
-

Reviewer #3 (Public review):
Summary:
The authors present a clearly written and beautifully presented piece of work demonstrating clear evidence to support the idea that BK channels and Cav1.3 channels can co-assemble prior to their assertion in the plasma membrane.
Strengths:
The experimental records shown back up their hypotheses and the authors are to be congratulated for the large number of control experiments shown in the ms.
-

Author response:
The following is the authors’ response to the original reviews.
Recommendations for the Authors:
(1) Clarify Mechanistic Interpretations
(a) Provide stronger evidence or a more cautious interpretation regarding whether intracellular BK-CaV1.3 ensembles are precursors to plasma membrane complexes.
This is an important point. We adjusted the interpretation regarding intracellular BKCaV1.3 hetero-clusters as precursors to plasma membrane complexes to reflect a more cautious stance, acknowledging the limitations of available data. We added the following to the manuscript.
“Our findings suggest that BK and CaV1.3 channels begin assembling intracellularly before reaching the plasma membrane, shaping their spatial organization and potentially facilitating functional coupling. While this suggests a coordinated process that …
Author response:
The following is the authors’ response to the original reviews.
Recommendations for the Authors:
(1) Clarify Mechanistic Interpretations
(a) Provide stronger evidence or a more cautious interpretation regarding whether intracellular BK-CaV1.3 ensembles are precursors to plasma membrane complexes.
This is an important point. We adjusted the interpretation regarding intracellular BKCaV1.3 hetero-clusters as precursors to plasma membrane complexes to reflect a more cautious stance, acknowledging the limitations of available data. We added the following to the manuscript.
“Our findings suggest that BK and CaV1.3 channels begin assembling intracellularly before reaching the plasma membrane, shaping their spatial organization and potentially facilitating functional coupling. While this suggests a coordinated process that may contribute to functional coupling, further investigation is needed to determine the extent to which these hetero-clusters persist upon membrane insertion.”
(b) Discuss the limitations of current data in establishing the proportion of intracellular complexes that persist on the cell surface.
We appreciate the suggestion. We expanded the discussion to address the limitations of current data in determining the proportion of intracellular complexes that persist on the cell surface. We added the following to the manuscript.
“Our findings highlight the intracellular assembly of BK-CaV1.3 hetero-clusters, though limitations in resolution and organelle-specific analysis prevent precise quantification of the proportion of intracellular complexes that ultimately persist on the cell surface. While our data confirms that hetero-clusters form before reaching the plasma membrane, it remains unclear whether all intracellular hetero-clusters transition intact to the membrane or undergo rearrangement or disassembly upon insertion. Future studies utilizing live cell tracking and high resolution imaging will be valuable in elucidating the fate and stability of these complexes after membrane insertion.”
(2) Refine mRNA Co-localization Analysis
(a) Include appropriate controls using additional transmembrane mRNAs to better assess the specificity of BK and CaV1.3 mRNA co-localization.
We agree with the reviewers that these controls are essential. We explain better the controls used to address this concern. We added the following to the manuscript.
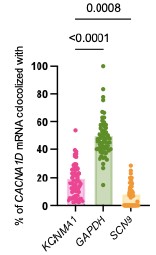
“To explore the origins of the initial association, we hypothesized that the two proteins are translated near each other, which could be detected as the colocalization of their mRNAs (Figure 5A and B). The experiment was designed to detect single mRNA molecules from INS-1 cells in culture. We performed multiplex in situ hybridization experiments using an RNAScope fluorescence detection kit to be able to image three mRNAs simultaneously in the same cell and acquired the images in a confocal microscope with high resolution. To rigorously assess the specificity of this potential mRNA-level organization, we used multiple internal controls. GAPDH mRNA, a highly expressed housekeeping gene with no known spatial coordination with channel mRNAs, served as a baseline control for nonspecific colocalization due to transcript abundance. To evaluate whether the spatial proximity between BK mRNA (KCNMA1) and CaV1.3 mRNA (CACNA1D) was unique to functionally coupled channels, we also tested for NaV1.7 mRNA (SCN9A), a transmembrane sodium channel expressed in INS-1 cells but not functionally associated with BK. This allowed us to determine whether the observed colocalization reflected a specific biological relationship rather than shared expression context. Finally, to test whether this proximity might extend to other calcium sources relevant to BK activation, we probed the mRNA of ryanodine receptor 2 (RyR2), another Ca2+ channel known to interact structurally with BK channels [32]. Together, these controls were chosen to distinguish specific mRNA colocalization patterns from random spatial proximity, shared subcellular distribution, or gene expression level artifacts.”
(b) Quantify mRNA co-localization in both directions (e.g., BK with CaV1.3 and vice versa) and account for differences in expression levels.
We thank the reviewer for this suggestion. We chose to quantify mRNA co-localization in the direction most relevant to the formation of functionally coupled hetero-clusters, namely, the proximity of BK (KCNMA1) mRNA to CaV1.3 (CACNA1D) mRNA. Since BK channel activation depends on calcium influx provided by nearby CaV1.3 channels, this directional analysis more directly informs the hypothesis of spatially coordinated translation and channel assembly. To address potential confounding effects of transcript abundance, we implemented a scrambled control approach in which the spatial coordinates of KCNMA1 mRNAs were randomized while preserving transcript count. This control resulted in significantly lower colocalization with CACNA1D mRNA, indicating that the observed proximity reflects a specific spatial association rather than expressiondriven overlap. We also assessed colocalization of CACNA1D with both KCNMA1, GAPDH mRNAs and SCN9 (NaV1.7); as you can see in the graph below these data support t the same conclusion but were not included in the manuscript.
Author response image 1.
(c) Consider using ER labeling as a spatial reference when analyzing mRNA localization
We thank the reviewers for this suggestion. Rather than using ER labeling as a spatial reference, we assess BK and CaV1.3 mRNA localization using fluorescence in situ hybridization (smFISH) alongside BK protein immunostaining. This approach directly identifies BK-associated translation sites, ensuring that observed mRNA localization corresponds to active BK synthesis rather than general ER association. By evaluating BK protein alongside its mRNA, we provide a more functionally relevant measure of spatial organization, allowing us to assess whether BK is synthesized in proximity to CaV1.3 mRNA within micro-translational complexes. The results added to the manuscript is as follows.
“To further investigate whether KCNMA1 and CACNA1D are localized in regions of active translation (Figure 7A), we performed RNAScope targeting KCNMA1 and CACNA1D alongside immunostaining for BK protein. This strategy enabled us to visualize transcript-protein colocalization in INS-1 cells with subcellular resolution. By directly evaluating sites of active BK translation, we aimed to determine whether newly synthesized BK protein colocalized with CACNA1D mRNA signals (Figure 7A). Confocal imaging revealed distinct micro-translational complex where KCNMA1 mRNA puncta overlapped with BK protein signals and were located adjacent to CACNA1D mRNA (Figure 7B). Quantitative analysis showed that 71 ± 3% of all KCNMA1 colocalized with BK protein signal which means that they are in active translation. Interestingly, 69 ± 3% of the KCNMA1 in active translation colocalized with CACNA1D (Figure 7C), supporting the existence of functional micro-translational complexes between BK and CaV1.3 channels.”
(3) Improve Terminology and Definitions
(a) Clarify and consistently use terms like "ensemble," "cluster," and "complex," especially in quantitative analyses.
We agree with the reviewers, and we clarified terminology such as 'ensemble,' 'cluster,' and 'complex' and used them consistently throughout the manuscript, particularly in quantitative analyses, to enhance precision and avoid ambiguity.
(b) Consider adopting standard nomenclature (e.g., "hetero-clusters") to avoid ambiguity.
We agree with the reviewers, and we adapted standard nomenclature, such as 'heteroclusters,' in the manuscript to improve clarity and reduce ambiguity.
(4) Enhance Quantitative and Image Analysis
(a) Clearly describe how colocalization and clustering were measured in super-resolution data.
We thank the reviewers for this suggestion. We have modified the Methods section to provide a clearer description of how colocalization and clustering were measured in our super-resolution data. Specifically, we now detail the image processing steps, including binary conversion, channel multiplication for colocalization assessment, and density-based segmentation for clustering analysis. These updates ensure transparency in our approach and improve accessibility for readers, and we added the following to the manuscript.
“Super-resolution imaging:
Direct stochastic optical reconstruction microscopy (dSTORM) images of BK and 1.3 overexpressed in tsA-201 cells were acquired using an ONI Nanoimager microscope equipped with a 100X oil immersion objective (1.4 NA), an XYZ closed-loop piezo 736 stage, and triple emission channels split at 488, 555, and 640 nm. Samples were imaged at 35°C. For singlemolecule localization microscopy, fixed and stained cells were imaged in GLOX imaging buffer containing 10 mM β-mercaptoethylamine (MEA), 0.56 mg/ml glucose oxidase, 34 μg/ml catalase, and 10% w/v glucose in Tris-HCl buffer. Single-molecule localizations were filtered using NImOS software (v.1.18.3, ONI). Localization maps were exported as TIFF images with a pixel size of 5 nm. Maps were further processed in ImageJ (NIH) by thresholding and binarization to isolate labeled structures. To assess colocalization between the signal from two proteins, binary images were multiplied. Particles smaller than 400 nm2 were excluded from the analysis to reflect the spatial resolution limit of STORM imaging (20 nm) and the average size of BK channels. To examine spatial localization preference, binary images of BK were progressively dilated to 20 nm, 40 nm, 60 nm, 80 nm, 100 nm, and 200 nm to expand their spatial representation. These modified images were then multiplied with the CaV1.3 channel to quantify colocalization and determine BK occupancy at increasing distances from CaV1.3. To ensure consistent comparisons across distance thresholds, data were normalized using the 200 nm measurement as the highest reference value, set to 1.”
(b) Where appropriate, quantify the proportion of total channels involved in ensembles within each compartment.
We thank the reviewers for this comment. However, our method does not allow for direct quantification of the total number of BK and CaV1.3 channels expressed within the ER or ER exit sites, as we rely on proximity-based detection rather than absolute fluorescence intensity measurements of individual channels. Traditional methods for counting total channel populations, such as immunostaining or single-molecule tracking, are not applicable to our approach due to the hetero-clusters formation process. Instead, we focused on the relative proportion of BK and CaV1.3 hetero-clusters within these compartments, as this provides meaningful insights into trafficking dynamics and spatial organization. By assessing where hetero-cluster preferentially localize rather than attempting to count total channel numbers, we can infer whether their assembly occurs before plasma membrane insertion. While this approach does not yield absolute quantification of ER-localized BK and CaV1.3 channels, it remains a robust method for investigating hetero-cluster formation and intracellular trafficking pathways. To reflect this limitation, we added the following to the manuscript.
“Finally, a key limitation of this approach is that we cannot quantify the proportion of total BK or CaV1.3 channels engaged in hetero-clusters within each compartment. The PLA method provides proximity-based detection, which reflects relative localization rather than absolute channel abundance within individual organelles”.
(5) Temper Overstated Claims
(a) Revise language that suggests the findings introduce a "new paradigm," instead emphasizing how this study extends existing models.
We agree with the reviewers, and we have revised the language to avoid implying a 'new paradigm.' The following is the significance statement.
“This work examines the proximity between BK and CaV1.3 molecules at the level of their mRNAs and newly synthesized proteins to reveal that these channels interact early in their biogenesis. Two cell models were used: a heterologous expression system to investigate the steps of protein trafficking and a pancreatic beta cell line to study the localization of endogenous channel mRNAs. Our findings show that BK and CaV1.3 channels begin assembling intracellularly before reaching the plasma membrane, revealing new aspects of their spatial organization. This intracellular assembly suggests a coordinated process that contributes to functional coupling.”
(b) Moderate conclusions where the supporting data are preliminary or correlative.
We agree with the reviewers, and we have moderated conclusions in instances where the supporting data are preliminary or correlative, ensuring a balanced interpretation. We added the following to the manuscript.
“This study provides novel insights into the organization of BK and CaV1.3 channels in heteroclusters, emphasizing their assembly within the ER, at ER exit sites, and within the Golgi. Our findings suggest that BK and CaV1.3 channels begin assembling intracellularly before reaching the plasma membrane, shaping their spatial organization, and potentially facilitating functional coupling. While this suggests a coordinated process that may contribute to functional coupling, further investigation is needed to determine the extent to which these hetero-clusters persist upon membrane insertion. While our study advances the understanding of BK and CaV1.3 heterocluster assembly, several key questions remain unanswered. What molecular machinery drives this colocalization at the mRNA and protein level? How do disruptions to complex assembly contribute to channelopathies and related diseases? Additionally, a deeper investigation into the role of RNA binding proteins in facilitating transcript association and localized translation is warranted”.
(6) Address Additional Technical and Presentation Issues
(a) Include clearer figure annotations, especially for identifying PLA puncta localization (e.g., membrane vs. intracellular).
We agree with the reviewers, and we have updated the figures to include clearer annotations that distinguish PLA puncta localized at the membrane versus those within intracellular compartments.
(b) Reconsider the scale and arrangement of image panels to better showcase the data.
We agree with the reviewers, and we have adjusted the scale and layout of the image panels to enhance data visualization and readability. Enlarged key regions now provide better clarity of critical features.
(c) Provide precise clone/variant information for BK and CaV1.3 channels used.
We thank the reviewers for their suggestion, and we now provide precise information regarding the BK and CaV1.3 channel constructs used in our experiments, including their Addgene plasmid numbers and relevant variant details. These have been incorporated into the Methods section to ensure reproducibility and transparency. We added the following to the manuscript.
“The CaV1.3 α subunit construct used in our study corresponds to the rat CaV1.3e splice variant containing exons 8a, 11, 31b, and 42a, with a deletion of exon 32. The BK channel construct used in this study corresponds to the VYR splice variant of the mouse BKα subunit (KCNMA1)”.
(d) Correct typographical errors and ensure proper figure/supplementary labeling throughout.
Typographical errors have been corrected, and figure/supplementary labeling has been reviewed for accuracy throughout the manuscript.
(7) Expand the Discussion
(a) Include a brief discussion of findings such as BK surface expression in the absence of CaV1.3.
We thank the reviewers for their suggestion. We expanded the Discussion to include a brief analysis of BK surface expression in the absence of CaV1.3. We included the following in the manuscript.
“BK Surface Expression and Independent Trafficking Pathways
BK surface expression in the absence of CaV1.3 indicates that its trafficking does not strictly rely on CaV1.3-mediated interactions. Since BK channels can be activated by multiple calcium sources, their presence in intracellular compartments suggests that their surface expression is governed by intrinsic trafficking mechanisms rather than direct calcium-dependent regulation. While some BK and CaV1.3 hetero-clusters assemble into signaling complexes intracellularly, other BK channels follow independent trafficking pathways, demonstrating that complex formation is not obligatory for all BK channels. Differences in their transport kinetics further reinforce the idea that their intracellular trafficking is regulated through distinct mechanisms. Studies have shown that BK channels can traffic independently of CaV1.3, relying on alternative calcium sources for activation [13, 41]. Additionally, CaV1.3 exhibits slower synthesis and trafficking kinetics than BK, emphasizing that their intracellular transport may not always be coordinated. These findings suggest that BK and CaV1.3 exhibit both independent and coordinated trafficking behaviors, influencing their spatial organization and functional interactions”.
(b) Clarify why certain colocalization comparisons (e.g., ER vs. ER exit sites) are not directly interpretable.
We thank the reviewer for their suggestion. A clarification has been added to the result section and discussion of the manuscript explaining why colocalization comparisons, such as ER versus ER exit sites, are not directly interpretable. We included the following in the manuscript.
“Result:
ER was not simply due to the extensive spatial coverage of ER labeling, we labeled ER exit sites using Sec16-GFP and probed for hetero-clusters with PLA. This approach enabled us to test whether the hetero-clusters were preferentially localized to ER exit sites, which are specialized trafficking hubs that mediate cargo selection and direct proteins from the ER into the secretory pathway. In contrast to the more expansive ER network, which supports protein synthesis and folding, ER exit sites ensure efficient and selective export of proteins to their target destinations”.
“By quantifying the proportion of BK and CaV1.3 hetero-clusters relative to total channel expression at ER exit sites, we found 28 ± 3% colocalization in tsA-201 cells and 11 ± 2% in INS-1 cells (Figure 3F). While the percentage of colocalization between hetero-clusters and the ER or ER exit sites alone cannot be directly compared to infer trafficking dynamics, these findings reinforce the conclusion that hetero-clusters reside within the ER and suggest that BK and CaV1.3 channels traffic together through the ER and exit in coordination”.
“Colocalization and Trafficking Dynamics
The colocalization of BK and CaV1.3 channels in the ER and at ER exit sites before reaching the Golgi suggests a coordinated trafficking mechanism that facilitates the formation of multi-channel complexes crucial for calcium signaling and membrane excitability [37, 38]. Given the distinct roles of these compartments, colocalization at the ER and ER exit sites may reflect transient proximity rather than stable interactions. Their presence in the Golgi further suggests that posttranslational modifications and additional assembly steps occur before plasma membrane transport, providing further insight into hetero-cluster maturation and sorting events. By examining BK-CaV1.3 hetero-cluster distribution across these trafficking compartments, we ensure that observed colocalization patterns are considered within a broader framework of intracellular transport mechanisms [39]. Previous studies indicate that ER exit sites exhibit variability in cargo retention and sorting efficiency [40], emphasizing the need for careful evaluation of colocalization data. Accounting for these complexities allows for a robust assessment of signaling complexes formation and trafficking pathways”.
Reviewer #1 (Recommendations for the authors):
In addition to the general aspects described in the public review, I list below a few points with the hope that they will help to improve the manuscript:
(1) Page 3: "they bind calcium delimited to the point of entry at calcium channels", better use "sources"
We agree with the reviewer. The phrasing on Page 3 has been updated to use 'sources' instead of 'the point of entry at calcium channels' for clarity.
(2) Page 3 "localized supplies of intracellular calcium", I do not like this term, but maybe this is just silly.
We agree with the reviewer. The term 'localized supplies of intracellular calcium' on Page 3 has been revised to “Localized calcium sources”
(3) Regarding the definitions stated by the authors: How do you distinguish between "ensembles" corresponding to "coordinated collection of BK and Cav channels" and "assembly of BK clusters with Cav clusters"? I believe that hetero-clusters is more adequate. The nomenclature does not respond to any consensus in the protein biology field, and I find that it introduces bias more than it helps. I would stick to heteroclusters nomenclature that has been used previously in the field. Moreover, in some discussion sections, the term "ensemble" is used in ways that border on vague, especially when talking about "functional signaling complexes" or "ensembles forming early." It's still acceptable within context but could benefit from clearer language to distinguish ensemble (structural proximity) from complex (functional consequence).
We agree with the reviewer, and we recognize the importance of precise nomenclature and have adopted hetero-clusters instead of ensembles to align with established conventions in the field. This term specifically refers to the spatial organization of BK and CaV1.3 channels, while functional complexes denote mechanistic interactions. We have revised sections where ensemble was used ambiguously to ensure clear distinction between structure and function.
The definition of "cluster" is clearly stated early but less emphasized in later quantitative analyses (e.g., particle size discussions in Figure 7). Figure 8 is equally confusing, graphs D and E referring to "BK ensembles" and "Cav ensembles", but "ensembles" should refer to combinations of both channels, whereas these seem to be "clusters". In fact, the Figure legend mentions "clusters".
We agree with the reviewer. Terminology has been revised throughout the manuscript to ensure consistency, with 'clusters' used appropriately in quantitative analyses and figure descriptions.
(4) Methods: how are clusters ("ensembles") analysed from the STORM data? What is the logarithm used for? More info about this is required. Equally, more information and discussion about how colocalization is measured and interpreted in superresolution microscopy are required.
We thank the reviewer for their suggestion, and additional details have been incorporated into the Methods section to clarify how clusters ('ensembles') are analyzed from STORM data, including the role of the logarithm in processing. Furthermore, we have expanded the discussion to provide more information on how colocalization is measured and interpreted in super resolution microscopy. We include the following in the manuscript.
“Direct stochastic optical reconstruction microscopy (dSTORM) images of BK and CaV1.3 overexpressed in tsA-201 cells were acquired using an ONI Nanoimager microscope equipped with a 100X oil immersion objective (1.4 NA), an XYZ closed-loop piezo 736 stage, and triple emission channels split at 488, 555, and 640 nm. Samples were imaged at 35°C. For singlemolecule localization microscopy, fixed and stained cells were imaged in GLOX imaging buffer containing 10 mM β-mercaptoethylamine (MEA), 0.56 mg/ml glucose oxidase, 34 μg/ml catalase, and 10% w/v glucose in Tris-HCl buffer. Single-molecule localizations were filtered using NImOS software (v.1.18.3, ONI). Localization maps were exported as TIFF images with a pixel size of 5 nm. Maps were further processed in ImageJ (NIH) by thresholding and binarization to isolate labeled structures. To assess colocalization between the signal from two proteins, binary images were multiplied. Particles smaller than 400 nm2 were excluded from the analysis to reflect the spatial resolution limit of STORM imaging (20 nm) and the average size of BK channels. To examine spatial localization preference, binary images of BK were progressively dilated to 20 nm, 40 nm, 60 nm, 80 nm, 100 nm, and 200 nm to expand their spatial representation. These modified images were then multiplied with the CaV1.3 channel to quantify colocalization and determine BK occupancy at increasing distances from CaV1.3. To ensure consistent comparisons across distance thresholds, data were normalized using the 200 nm measurement as the highest reference value, set to 1”.
(5) Related to Figure 2:
(a) Why use an antibody to label GFP when PH-PLCdelta should be a membrane marker? Where is the GFP in PH-PKC-delta (intracellular, extracellular? Images in Figure 2E are confusing, there is a green intracellular signal.
We thank the reviewer for their feedback. To clarify, GFP is fused to the N-terminus of PH-PLCδ and primarily localizes to the inner plasma membrane via PIP2 binding. Residual intracellular GFP signal may reflect non-membrane-bound fractions or background from anti-GFP immunostaining. We added a paragraph explaining the use of the antibody anti GFP in the Methods section Proximity ligation assay subsection.
(b) The images in Figure 2 do not help to understand how the authors select the PLA puncta located at the plasma membrane. How do the authors do this? A useful solution would be to indicate in Figure 2 an example of the PLA signals that are considered "membrane signals" compared to another example with "intracellular signals". Perhaps this was intended with the current Figure, but it is not clear.
We agree with the reviewer. We have added a sentence to explain how the number of PLA puncta at the plasma membrane was calculated.
“We visualized the plasma membrane with a biological sensor tagged with GFP (PHPLCδ-GFP) and then probed it with an antibody against GFP (Figure 2E). By analyzing the GFP signal, we created a mask that represented the plasma membrane. The mask served to distinguish between the PLA puncta located inside the cell and those at the plasma membrane, allowing us to calculate the number of PLA puncta at the plasma membrane”.
(c) Figure 2C: What is the negative control? Apologies if it is described somewhere, but I seem not to find it in the manuscript.
We thank the reviewer for their suggestion. For the negative control in Figure 2C, BK was probed using the primary antibody without co-staining for CaV1.3 or other proteins, ensuring specificity and ruling out non-specific antibody binding or background fluorescence. A sentence clarifying the negative control for Figure 2C has been added to the Results section, specifying that BK was probed using the primary antibody without costaining for CaV1.3 or other proteins to ensure specificity.
“To confirm specificity, a negative control was performed by probing only for BK using the primary antibody, ensuring that detected signals were not due to non-specific binding or background fluorescence”.
(d) What is the resolution in z of the images shown in Figure 2? This is relevant for the interpretation of signal localization.
The z-resolution of the images shown in Figure 2 was approximately 270–300 nm, based on the Zeiss Airyscan system’s axial resolution capabilities. Imaging was performed with a step size of 300 nm, ensuring adequate sampling for signal localization while maintaining optimal axial resolution.
“In a different experiment, we analyzed the puncta density for each focal plane of the cell (step size of 300 nm) and compared the puncta at the plasma membrane to the rest of the cell”.
(e) % of total puncta in PM vs inside cell are shown for transfected cells, what is this proportion in INS-1 cells?
This quantification was performed for transfected cells; however, we have not conducted the same analysis in INS-1 cells. Future experiments could address this to determine potential differences in puncta distribution between endogenous and overexpressed conditions.
(6) Related to Figure 3:
(a) Figure 3B: is this antibody labelling or GFP fluorescence? Why do they use GFP antibody labelling, if the marker already has its own fluorescence? This should at least be commented on in the manuscript.
We thank the reviewer for their concern. In Figure 3B, GFP was labeled using an antibody rather than relying on its intrinsic fluorescence. This approach was necessary because GFP fluorescence does not withstand the PLA protocol, resulting in significant fading. Antibody labeling provided stronger signal intensity and improved resolution, ensuring optimal signal-to-noise ratio for accurate analysis.
A clarification regarding the use of GFP antibody labeling in Figure 3B has been added to the Methods section, explaining that intrinsic GFP fluorescence does not endure the PLA protocol, necessitating antibody-based detection for improved signal and resolution.We added the following to the manuscript.
“For PLA combined with immunostaining, PLA was followed by a secondary antibody incubation with Alexa Fluor-488 at 2 μg/ml for 1 hour at 21˚C. Since GFP fluorescence fades significantly during the PLA protocol, resulting in reduced signal intensity and poor image resolution, GFP was labeled using an antibody rather than relying on its intrinsic fluorescence”.
(b) Why is it relevant to study the ER exit sites? Some explanation should be included in the main text (page 11) for clarification to non-specialized readers. Again, the quantification should be performed on the proportion of clusters/ensembles out of the total number of channels expressed at the ER (or ER exit sites).
We thank the reviewer for their feedback. We have modified this section to include a more detailed explanation of the relevance of ER exit sites to protein trafficking. ER exit sites serve as specialized sorting hubs that regulate the transition of proteins from the ER to the secretory pathway, distinguishing them from the broader ER network, which primarily facilitates protein synthesis and folding. This additional context clarifies why studying ER exit sites provides valuable insights into ensemble trafficking dynamics.
Regarding quantification, our method does not allow for direct measurement of the total number of BK and CaV1.3 channels expressed at the ER or ER exit sites. Instead, we focused on the proportion of hetero-clusters localized within these compartments, which provides insight into trafficking pathways despite the limitation in absolute channel quantification. We included the following in the manuscript in the Results section.
“To determine whether the observed colocalization between BK–CaV1.3 hetero-clusters and the ER was not simply due to the extensive spatial coverage of ER labeling, we labeled ER exit sites using Sec16-GFP and probed for hetero-clusters with PLA. This approach enabled us to test whether the hetero-clusters were preferentially localized to ER exit sites, which are specialized trafficking hubs that mediate cargo selection and direct proteins from the ER into the secretory pathway. In contrast to the more expansive ER network, which supports protein synthesis and folding, ER exit sites ensure efficient and selective export of proteins to their target destinations”.
“By quantifying the proportion of BK and CaV1.3 hetero-clusters relative to total channel expression at ER exit sites, we found 28 ± 3% colocalization in tsA-201 cells and 11 ± 2% in INS-1 cells (Figure 3F). While the percentage of colocalization between hetero-clusters and the ER or ER exit sites alone cannot be directly compared to infer trafficking dynamics, these findings reinforce the conclusion that hetero-clusters reside within the ER and suggest that BK and CaV1.3 channels traffic together through the ER and exit in coordination”.
(7) Related to Figure 4:
A control is included to confirm that the formation of BK-Cav1.3 ensembles is not unspecific. Association with a protein from the Golgi (58K) is tested. Why is this control only done for Golgi? No similar experiment has been performed in the ER. This aspect should be commented on.
We thank the reviewer for their suggestion. We selected the Golgi as a control because it represents the final stage of protein trafficking before proteins reach their functional destinations. If BK and CaV1.3 hetero-cluster formation is specific at the Golgi, this suggests that their interaction is maintained throughout earlier trafficking steps, including within the ER. While we did not perform an equivalent control experiment in the ER, the Golgi serves as an effective checkpoint for evaluating specificity within the broader protein transport pathway. We included the following in the manuscript.
“We selected the Golgi as a control because it represents the final stage of protein trafficking, ensuring that hetero-cluster interactions observed at this point reflect specificity maintained throughout earlier trafficking steps, including within the ER”.
(8) How is colocalization measured, eg, in Figure 6? Are the images shown in Figure 6 representative? This aspect would benefit from a clearer description.
We thank the reviewer for their suggestion. A section clarifying colocalization measurement and the representativeness of Figure 6 images has been added to the Methods under Data Analysis. We included the following in the manuscript.
For PLA and RNAscope experiments, we used custom-made macros written in ImageJ. Processing of PLA data included background subtraction. To assess colocalization, fluorescent signals were converted into binary images, and channels were multiplied to identify spatial overlap.
(9) The text should be revised for typographical errors, for example:
(a) Summary "evidence of" (CHECK THIS ONE)
We agree with the reviewer, and we corrected the typographical errors
(b) Table 1, row 3: "enriches" should be "enrich"
We agree with the reviewer. The term 'enriches' in Table 1, row 3 has been corrected to 'enrich'.
(c) Figure 2B "priximity"
We agree with the reviewer. The typographical errors in Figure 2B has been corrected from 'priximity' to 'proximity'.
(d) Legend of Figure 7 (C) "size of BK and Cav1.3 channels". Does this correspond to individual channels or clusters?
We agree with the reviewer. The legend of Figure 7C has been clarified to indicate that 'size of BK and Cav1.3 channels' refers to clusters rather than individual channels.
(e) Methods: In the RNASCOPE section, "Fig.4-supp1" should be "Fig. 5-supp1"
(f) Page 15, Figure 5B is cited, should be Figure 6B
We agree with the reviewer. The reference in the RNASCOPE section has been updated from 'Fig.4-supp1' to 'Fig. 5-supp1,' and the citation on Page 15 has been corrected from Figure 5B to Figure 6B.
Reviewer #2 (Recommendations for the authors):
(1) The abstract could be more accessible for a wider readership with improved flow.
We thank the reviewer for their suggestion. We modified the summary as follows to provide a more coherent flow for a wider readership.
“Calcium binding to BK channels lowers BK activation threshold, substantiating functional coupling with calcium-permeable channels. This coupling requires close proximity between different channel types, and the formation of BK–CaV1.3 hetero-clusters at nanometer distances exemplifies this unique organization. To investigate the structural basis of this interaction, we tested the hypothesis that BK and CaV1.3 channels assemble before their insertion into the plasma membrane. Our approach incorporated four strategies: (1) detecting interactions between BK and CaV1.3 proteins inside the cell, (2) identifying membrane compartments where intracellular hetero-clusters reside, (3) measuring the proximity of their mRNAs, and (4) assessing protein interactions at the plasma membrane during early translation. These analyses revealed that a subset of BK and CaV1.3 transcripts are spatially close in micro-translational complexes, and their newly synthesized proteins associate within the endoplasmic reticulum (ER) and Golgi. Comparisons with other proteins, transcripts, and randomized localization models support the conclusion that BK and CaV1.3 hetero-clusters form before their insertion at the plasma membrane”.
(2) Figure 2B - spelling of proximity.
We agree with the reviewer. The typographical errors in Figure 2B has been corrected from 'priximity' to 'proximity'.
Reviewer #3 (Recommendations for the authors):
Minor issues to improve the manuscript:
(1) For completeness, the authors should include a few sentences and appropriate references in the Introduction to mention that BK channels are regulated by auxiliary subunits.
We agree with the reviewer. We have revised the Introduction to include a brief discussion of how BK channel function is modulated by auxiliary subunits and provided appropriate references to ensure completeness. These additions highlight the broader regulatory mechanisms governing BK channel activity, complementing the focus of our study. We included the following in the manuscript.
“Additionally, BK channels are modulated by auxiliary subunits, which fine-tune BK channel gating properties to adapt to different physiological conditions. β and γ subunits regulate BK channel kinetics, altering voltage sensitivity and calcium responsiveness [18]. These interactions ensure precise control over channel activity, allowing BK channels to integrate voltage and calcium signals dynamically in various cell types. Here, we focus on the selective assembly of BK channels with CaV1.3 and do not evaluate the contributions of auxiliary subunits to BK channel organization.”
(2) Insert a space between 'homeostasis' and the square bracket at the end of the Introduction's second paragraph.
We agree with the reviewer. A space has been inserted between 'homeostasis' and the square bracket in the second paragraph of the Introduction for clarity.
(3) The images presented in Figures 2-5 should be increased in size (if permitted by the Journal) to allow the reader to clearly see the puncta in the fluorescent images. This would necessitate reconfiguring the figures into perhaps a full A4 page per figure, but I think the quality of the images presented really do deserve to "be seen". For example, Panels A & B could be at the top of Figure 2, with C & D presented below them. However, I'll leave it up to the authors to decide on the most aesthetically pleasing way to show these.
We agree with the reviewer. We have increased the size of Figures 2–8 to enhance the visibility of fluorescent puncta, as suggested. To accommodate this, we reorganized the panel layout for each figure—for example, in Figure 2, Panels A and B are now placed above Panels C and D to support a more intuitive and aesthetically coherent presentation. We believe this revised configuration highlights the image quality and improves readability while conforming to journal layout constraints.
(4) I think that some of the sentences could be "toned down"
(a) eg, in the first paragraph below Figure 2, the authors state "that 46(plus minus)3% of the puncta were localised on intracellular membranes" when, at that stage, no data had been presented to confirm this. I think changing it to "that 46(plus minus)3% of the puncta were localised intracellularly" would be more precise.
(b) Similarly, please consider replacing the wording of "get together at membranes inside the cell" to "co-localise intracellularly".
(c) In the paragraph just before Figure 5, the authors mention that "the abundance of KCNMA1 correlated more with the abundance of CACNA1D than ... with GAPDH." Although this is technically correct, the R2 value was 0.22, which is exceptionally poor. I don't think that the paper is strengthened by sentences such as this, and perhaps the authors might tone this down to reflect this.
(d) The authors clearly demonstrate in Figure 8 that a significant number of BK channels can traffic to the membrane in the absence of Cav1.3. Irrespective of the differences in transcription/trafficking time between the two channel types, the authors should insert a few lines into their discussion to take this finding into account.
We appreciate the reviewer’s feedback regarding the clarity and precision of our phrasing.
Our responses for each point are below.
(a) We have modified the statement in the first paragraph below Figure 2, changing '46 ± 3% of the puncta were localized on intracellular membranes' to '46 ± 3% of the puncta were localized ‘intracellularly’ to ensure accuracy in the absence of explicit data confirming membrane association.
(b) Similarly, we have replaced 'get together at membranes inside the cell' with 'colocalize intracellularly' to maintain clarity and avoid unintended implications.
(c) Regarding the correlation between KCNMA1 and CACNA1D abundance, we recognize that the R² value of 0.22 is relatively low. To reflect this appropriately, we have revised the phrasing to indicate that while a correlation exists, it is modest. We added the following to the manuscript.
“Interestingly, the abundance of KCNMA1 transcripts correlated more with the abundance of CACNA1D transcripts than with the abundance of GAPDH, a standard housekeeping gene, though with a modest R² value.”
(d) To incorporate the findings from Figure 8, we have added discussion acknowledging that a substantial number of BK channels traffic to the membrane independently of CaV1.3. This addition provides context for potential trafficking mechanisms that operate separately from ensemble formation.
(5) For clarity, please insert the word "total" in the paragraph after Figure 3 "..."63{plus minus}3% versus 50%{plus minus}6% of total PLA puncta were localised at the ER". I know this is explicitly stated later in the manuscript, but I think it needs to be clarified earlier.
We agree with the reviewer. The word 'total' has been inserted in the paragraph following Figure 3 to clarify the percentage of PLA puncta localized at the ER earlier in the manuscript
(6) In the discussion, I think an additional (short) paragraph needs to be included to clarify to the reader why the % "colocalization between ensembles and the ER or the ER exit sites can't be compared or used to understand the dynamics of the ensembles". This may permit the authors to remove the last sentence of the paragraph just before the results section, "BK and Cav1.3 ensembles go through the Golgi."
We thank the reviewer for their suggestion. We have added a short paragraph in the discussion to clarify why colocalization percentages between ensembles and the ER or ER exit sites cannot be compared to infer ensemble dynamics. This allowed us to remove the final sentence of the paragraph preceding the results section ('BK and Cav1.3 ensembles go through the Golgi).
(7) In the paragraph after Figure 6, Figure 5B is inadvertently referred to. Please correct this to Figure 6B.
We agree with the reviewer. The reference to Figure 5B in the paragraph after Figure 6 has been corrected to Figure 6B.
(8) In the discussion under "mRNA co-localisation and Protein Trafficking", please insert a relevant reference illustrating that "disruption in mRNA localization... can lead to ion channel mislocalization".
We agree with the reviewer. We have inserted a relevant reference under 'mRNA Colocalization and Protein Trafficking' to illustrate that disruption in mRNA localization can lead to ion channel mislocalization.
(9) The supplementary Figures appear to be incorrectly numbered. Please correct and also ensure that they are correctly referred to in the text.
We agree with the reviewer. The numbering of the supplementary figures has been corrected, and all references to them in the text have been updated accordingly.
(10) The final panels of the currently labelled Figure 5-Supplementary 2 need to have labels A-F included on the image.
We agree with the reviewer. Labels A-F have been added to the final panels of Figure 5-Supplementary 2.
References
(1) Shah, K.R., X. Guan, and J. Yan, Structural and Functional Coupling of Calcium-Activated BK Channels and Calcium-Permeable Channels Within Nanodomain Signaling Complexes. Frontiers in Physiology, 2022. Volume 12 - 2021.
(2) Chen, A.L., et al., Calcium-Activated Big-Conductance (BK) Potassium Channels Traffic through Nuclear Envelopes into Kinocilia in Ray Electrosensory Cells. Cells, 2023. 12(17): p. 2125.
(3) Berkefeld, H., B. Fakler, and U. Schulte, Ca2+-activated K+ channels: from protein complexes to function. Physiol Rev, 2010. 90(4): p. 1437-59.
(4) Loane, D.J., P.A. Lima, and N.V. Marrion, Co-assembly of N-type Ca2+ and BK channels underlies functional coupling in rat brain. J Cell Sci, 2007. 120(Pt 6): p. 98595.
(5) Boncompain, G. and F. Perez, The many routes of Golgi-dependent trafficking. Histochemistry and Cell Biology, 2013. 140(3): p. 251-260.
(6) Kurokawa, K. and A. Nakano, The ER exit sites are specialized ER zones for the transport of cargo proteins from the ER to the Golgi apparatus. The Journal of Biochemistry, 2019. 165(2): p. 109-114.
(7) Chen, G., et al., BK channel modulation by positively charged peptides and auxiliary γ subunits mediated by the Ca2+-bowl site. Journal of General Physiology, 2023. 155(6).
-

-

Author Response:
We thank the reviewers for their thoughtful feedback and appreciate their recognition of the value of our findings. In response, we are refining the manuscript to clarify key terminology, more clearly describe our image analysis workflows, and temper the interpretation of our results where appropriate. We are planning to perform additional experiments to further investigate the specificity of mRNA co-localization between BK and CaV1.3 channels. We acknowledge the importance of understanding ensemble trafficking dynamics and the functional role of pre-assembly at the plasma membrane, and we plan to explore these questions in future work. We look forward to submitting a revised manuscript that addresses the reviewers’ comments in detail.
-

eLife Assessment
This valuable manuscript provides convincing evidence that BK and CaV1.3 channels can co-localize as ensembles early in the biosynthetic pathway, including in the ER and Golgi. The findings, supported by a range of imaging and proximity assays, offer insights into channel organization in both heterologous and endogenous systems. However, mechanistic questions remain unresolved, particularly regarding the specificity of mRNA co-localization, the dynamics of ensemble trafficking, and the functional significance of pre-assembly at the plasma membrane. While the data broadly support the central claims, certain conclusions would benefit from more restrained interpretation and additional clarification to enhance the manuscript's impact and rigor.
-

Joint Public Review:
This study presents a valuable contribution to our understanding of ion channel complex assembly by investigating whether BK and CaV1.3 channels begin to form functional associations early in the biosynthetic pathway, prior to reaching the plasma membrane. Using a combination of proximity ligation assays, single-molecule RNA imaging, and super-resolution microscopy, the authors provide convincing evidence that these channels co-localize intracellularly within the ER and Golgi, in both overexpression systems and a relevant endogenous cell model. The study addresses an important and underexplored aspect of membrane protein trafficking and organization, with broader implications for how ion channel signaling complexes are assembled and regulated. The experimental approaches are generally appropriate and the imaging …
Joint Public Review:
This study presents a valuable contribution to our understanding of ion channel complex assembly by investigating whether BK and CaV1.3 channels begin to form functional associations early in the biosynthetic pathway, prior to reaching the plasma membrane. Using a combination of proximity ligation assays, single-molecule RNA imaging, and super-resolution microscopy, the authors provide convincing evidence that these channels co-localize intracellularly within the ER and Golgi, in both overexpression systems and a relevant endogenous cell model. The study addresses an important and underexplored aspect of membrane protein trafficking and organization, with broader implications for how ion channel signaling complexes are assembled and regulated. The experimental approaches are generally appropriate and the imaging data are clearly presented, with a commendable number of control experiments included. However, several limitations temper the interpretation of the results. The mechanisms underlying mRNA co-localization, and the role of co-translation in complex formation, remain insufficiently defined. Similarly, while intracellular colocalization is convincingly demonstrated, the study does not establish whether such early assembly is the predominant pathway for generating functional complexes at the plasma membrane. More rigorous quantification of channel co-association across compartments, and clarification of key terminology and image analysis methods, would strengthen the overall conclusions. Some of the language in the manuscript would also benefit from a more measured tone to avoid overstating the novelty of the findings. Despite these limitations, the study offers meaningful insights into intracellular ion channel organization and will be of interest to researchers in cell biology, membrane trafficking, and neurophysiology. With focused revisions addressing the outlined points, the manuscript has the potential to make a solid contribution to the field.
-

-