The mechanism of DRB7.2:DRB4 mediated sequestering of endogenous inverted-repeat dsRNA precursors in plants
Curation statements for this article:-
Curated by eLife
eLife Assessment
The manuscript provides valuable findings in the field for understanding the RNAi regulation in plants at the molecular level with a model of how DRB7.2 and DRB4 form a heterodimer and protect dsRNA from DICER activity. The presented data provide a solid basis for the model, but certain measurements could benefit from replicates for robust statistics.
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Abstract
Noncoding transcribable inverted repeat sequences are vital in genome stability, regulation of transposable elements, mutations, and diseases in eukaryotes. In vascular plants, dsRNA Binding Proteins (dsRBPs), DRB7.2 and DRB4, inhibit Dicer-like-protein 3 (DCL3) to stall endogenous inverted-repeat dsRNA (endo-IR dsRNA) mediated gene silencing. As dsRBPs generally assist Dicers, the inhibition of DCL3 by a dsRBP complex is quite enigmatic. Here, we explore how the DRB7.2:DRB4 complex sequesters substrate dsRNA of DCL3 using a structure-based mechanistic approach. Intriguingly, the crucial step of endo-IR dsRNA precursor sequestration is the high affinity complex formation of interacting domains of DRB7.2 (DRB7.2M) and DRB4 (DRB4D3). Next, we establish that DRB7.2 simultaneously interacts with DRB4 and endo-IR dsRNA precursors, where DRB4 contributes towards enhancement in the affinity of the complex with dsRNA, thereby impairing DCL3 mediated cleavage of endo-IR dsRNA precursors. The uniqueness of the DRB4D3 structure implies that the trans-acting (tasi)/siRNA initiation complex formed by DCL4:DRB4 in plants is diverse from its non-plant higher eukaryotes. Overall, we present considerable insights into endo-IR dsRNA precursors regulation in plants and indicate a differential evolution of RNAi initiation complexes between plants and other higher eukaryotes.
Article activity feed
-

eLife Assessment
The manuscript provides valuable findings in the field for understanding the RNAi regulation in plants at the molecular level with a model of how DRB7.2 and DRB4 form a heterodimer and protect dsRNA from DICER activity. The presented data provide a solid basis for the model, but certain measurements could benefit from replicates for robust statistics.
-

Reviewer #1 (Public review):
Summary:
In this manuscript, Paturi et.al. presents a detailed structural and mechanistic study of the DRB7.2:DRB4 complex in plants, focusing on its role in sequestering endogenous inverted-repeat dsRNA precursors and inhibiting Dicer-like protein 3 (DCL3) activity. By truncating the two proteins, they systematically identify the domains involved in direct interaction between DRB7.2 and DRB4 and study the interactions between the two using biophysical techniques (ITC and NMR). They show using NMR that the interacting domains between the two proteins are likely partially unfolded or aggregated in the absence of the binding partner and determining the NMR structure of the individual interacting domains in the presence of the isotopically unlabelled partner using sparse restrain data combined with Rosetta. …
Reviewer #1 (Public review):
Summary:
In this manuscript, Paturi et.al. presents a detailed structural and mechanistic study of the DRB7.2:DRB4 complex in plants, focusing on its role in sequestering endogenous inverted-repeat dsRNA precursors and inhibiting Dicer-like protein 3 (DCL3) activity. By truncating the two proteins, they systematically identify the domains involved in direct interaction between DRB7.2 and DRB4 and study the interactions between the two using biophysical techniques (ITC and NMR). They show using NMR that the interacting domains between the two proteins are likely partially unfolded or aggregated in the absence of the binding partner and determining the NMR structure of the individual interacting domains in the presence of the isotopically unlabelled partner using sparse restrain data combined with Rosetta. They also determine the complex structure of the interacting DRB7.2 dsRBD domain and the DRB4 D3 domain using X-ray crystallography.
Strengths:
Overall, the manuscript is well written, provides molecular details at high resolution between the interaction of DRB7.2 and DRB4 and the data in the manuscript strongly supports the proposed model where DRB7.2:DRB4 complex sequesters the DCL3 substrates inhibiting its function of producing epigenetically activated siRNAs.
Weaknesses:
Major comments:
(1) The manuscript unfortunately completely lacks functional validation of the determined DRB7.2:DRB4 complex structure which is required for the rigorous validation of the proposed model. For functional validation of the determined structures, the author should at least present the mutational analysis (impact on complex formation, RNA affinity) of the point mutants derived from the structure of the DRB7.2:DRB4 complex.
(2) The proposed model shows the DRB7.2M and DRB4D3 as partially folded/aggregated proteins in the absence of the complex, understandably from the presented NMR data of the individual domains. However, in the cellular context, when the RNAs are present, especially DRB7.2M might be properly folded/not aggregated. Could the authors support or negate this by showing the 15N HSQC spectrum of DRB7.2M in complex with the 13 bp dsRNA?
(3) It remains unclear from the manuscript if DRB7.1 will have a similar or different mechanism of interaction with DRB4. Based on the sequence comparisons of the two proteins, the authors should comment on this in the discussion section.
Minor comments:
(1) There are no errors for the N, dH and dS values of the ITC measurements in Table 1. Also, it seems that the measurements are done only once. Values derived from at least triplicates should be presented. This would be helpful to increase confidence in the values derived from ITC especially for the titration between DRB7.2, DRB4C, and DRB4D3 as the N value there is substantially lower than 1 which does not agree with the other data.
-

Reviewer #2 (Public review):
Summary:
The manuscript by Paturi and colleagues uses an approach that combines structural biology and biochemistry to probe protein-protein and protein-RNA interactions for two protein factors related to the dsRNA pathway in plants.
Strengths:
A key finding in the research is the direct demonstration of the ability of the single dsRBD (double-strand RNA binding domain) of DRB7.2 to interact simultaneously with dsRNA as well as the C-terminal domain of DRB4. The heterodimerization of DRB7.2 and DRB4 is demonstrated to make a high-affinity complex with dsRNA and it is proposed that this atypical use of the dsRBD domain to bridge the protein and RNA may contribute to the ability to prevent cleavage that would otherwise occur for dsRNA. The primary results for the interactions are generally well-supported by …
Reviewer #2 (Public review):
Summary:
The manuscript by Paturi and colleagues uses an approach that combines structural biology and biochemistry to probe protein-protein and protein-RNA interactions for two protein factors related to the dsRNA pathway in plants.
Strengths:
A key finding in the research is the direct demonstration of the ability of the single dsRBD (double-strand RNA binding domain) of DRB7.2 to interact simultaneously with dsRNA as well as the C-terminal domain of DRB4. The heterodimerization of DRB7.2 and DRB4 is demonstrated to make a high-affinity complex with dsRNA and it is proposed that this atypical use of the dsRBD domain to bridge the protein and RNA may contribute to the ability to prevent cleavage that would otherwise occur for dsRNA. The primary results for the interactions are generally well-supported by the data, and the conclusions are taken from the available results without excessive speculation.
Weaknesses:
There is a need for some statistical repeats, as well as a suggested movement of many protein characterization findings in the solution state to support data or to better indicate how these properties could play a role in the final proposed mechanism. There is also the need for certain measurement replicates, such as for the ITC data which are derived from single measurements and lack sufficient estimates of error.
-

Author response:
Public Reviews:
Reviewer #1 (Public review):
Summary:
In this manuscript, Paturi et.al. presents a detailed structural and mechanistic study of the DRB7.2:DRB4 complex in plants, focusing on its role in sequestering endogenous inverted-repeat dsRNA precursors and inhibiting Dicer-like protein 3 (DCL3) activity. By truncating the two proteins, they systematically identify the domains involved in direct interaction between DRB7.2 and DRB4 and study the interactions between the two using biophysical techniques (ITC and NMR). They show using NMR that the interacting domains between the two proteins are likely partially unfolded or aggregated in the absence of the binding partner and determining the NMR structure of the individual interacting domains in the presence of the isotopically unlabelled partner using sparse …
Author response:
Public Reviews:
Reviewer #1 (Public review):
Summary:
In this manuscript, Paturi et.al. presents a detailed structural and mechanistic study of the DRB7.2:DRB4 complex in plants, focusing on its role in sequestering endogenous inverted-repeat dsRNA precursors and inhibiting Dicer-like protein 3 (DCL3) activity. By truncating the two proteins, they systematically identify the domains involved in direct interaction between DRB7.2 and DRB4 and study the interactions between the two using biophysical techniques (ITC and NMR). They show using NMR that the interacting domains between the two proteins are likely partially unfolded or aggregated in the absence of the binding partner and determining the NMR structure of the individual interacting domains in the presence of the isotopically unlabelled partner using sparse restrain data combined with Rosetta. They also determine the complex structure of the interacting DRB7.2 dsRBD domain and the DRB4 D3 domain using X-ray crystallography.
Strengths:
Overall, the manuscript is well written, provides molecular details at high resolution between the interaction of DRB7.2 and DRB4, and the data in the manuscript strongly supports the proposed model where DRB7.2:DRB4 complex sequesters the DCL3 substrates inhibiting its function of producing epigenetically activated siRNAs.
Weaknesses:
Major comments:
(1) The manuscript, unfortunately, completely lacks functional validation of the determined DRB7.2:DRB4 complex structure, which is required for the rigorous validation of the proposed model. For functional validation of the determined structures, the author should at least present the mutational analysis (impact on complex formation, RNA affinity) of the point mutants derived from the structure of the DRB7.2:DRB4 complex.
We thank the reviewer for pointing out a crucial aspect that is missed out in our manuscript. With the inputs and experiments proposed above, we would certainly like to perform additional mutational analysis to determine the impact on the heterodimeric complex formation and identify the key essential residues involved in the RNA binding.
We expect that we can accomplish this study in the next ~ 4-6 months as we may have to create a combination of mutations for residues involved in the dimerization interface, namely, T131, V132, E134, F136, W156, and V161 on DRB7.2M. Having said that, the disruption of the heterodimer interface would probably lead to DRB7.2M and DRB4D3 returning to their fast-intermediate timescale exchanging native homo-oligomeric state/partially folded state.
For dsRNA binding, six residues (i.e., A85 and K86 (a1), H112 and K114 (b1-b2 loop), and K142 and K144 (a2)) involved in the RNA binding interface and a few other residues based on the mutational data will be considered.
(2) The proposed model shows the DRB7.2M and DRB4D3 as partially folded/aggregated proteins in the absence of the complex, understandably from the presented NMR data of the individual domains. However, in the cellular context, when the RNAs are present, especially DRB7.2M might be properly folded/not aggregated. Could the authors support or negate this by showing the 15N HSQC spectrum of DRB7.2M in complex with the 13 bp dsRNA?
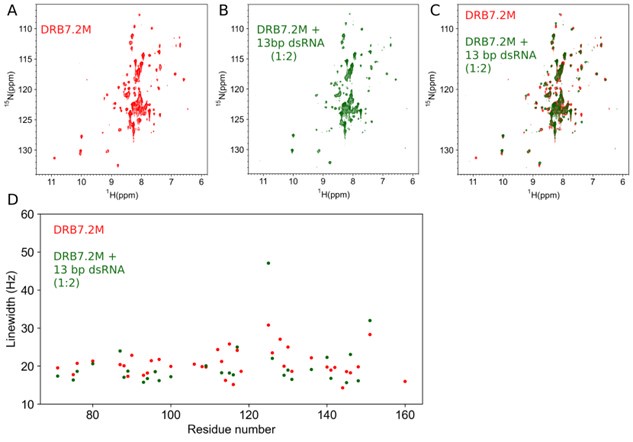
While we have no direct proof that the DRB7.2M might be folded/not aggregated in the presence of RNAs in the cellular context, the in vitro NMR-based titration studies of alone DRB7.2 (Author response image 1A) with two molar equivalence of 13 bp dsRNA (Author response image 1B and R1C) indicate that there is no change in overall spectral pattern (except for the apparent chemical shift perturbations as expected from fast-intermediate exchange timescale binding of DRB7.2M with 13 bp dsRNA), implying that the dsRNA alone is neither necessary nor sufficient to disrupt the native fast exchange oligomeric states sampled by individual DRB7.2 and DRB7.2M.
Author response image 1.
DRB7.2M binding interaction with 13bp dsRNA (A) 1H-15N TROSY-HSQC of U[15N, 2H] DRB7.2M. (B) 1H-15N TROSY-HSQC of U[15N, 2H] DRB7.2M in the presence of 13 bp dsRNA with 1:2 molar equivalence. (C) An overlay of (A) and (B) indicates no evident changes in the broadening of resonances. (D) The 15N linewidth analysis of unbound (red) and bound (green) forms of U[15N, 2H] DRB7.2M resonances for which the assignment could be traced from the assignments of the DRB7.2M:DRB4D3 complex.
Furthermore, the line-width analysis, shown in Author response image 1D, implies that the ~R2 rates are roughly identical in the presence of dsRNA, indicating that the native oligomeric state of DRB7.2M remains unperturbed by the presence of dsRNA. Our observation also corroborates with the crystal structure presented in the manuscript, where we have observed that the hetero-dimeric interface lies on the opposite side of the dsRNA binding interface of the DRB7.2M:DRB4D3 complex.
Therefore, the dsRNA substrate does not have any role in the native partially folded/oligomeric state of DRB7.2M.
(3) It remains unclear from the manuscript if DRB7.1 will have a similar or different mechanism of interaction with DRB4. Based on the sequence comparisons of the two proteins, the authors should comment on this in the discussion section.
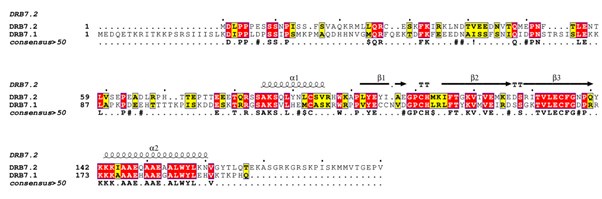
Pairwise sequence alignment of full-length DRB7.2 and DRB7.1 reveals 50.7% similarity and a 33.2% identity derived from EMBOSS Needle (Author response image 2).
Author response image 2.
ClustalW alignment of full-length DRB7.2 and DRB7.1. The secondary structure elements are derived from the crystal structure of DRB7.2M (PDB ID: 8IGD). Identical residues are marked with red highlights, whereas similar residues are marked with yellow highlights, and the consensus residues (> 50%) are annotated below the sequence alignment.
As expected, for the dsRBD region (corresponding to DRB7.2M), we observe a much higher degree of alignment with a 76.7% similarity with a 54.7% identity (Author response image 3).
Author response image 3.
ClustalW alignment of the dsRBD region of DRB7.2 and DRB7.1. The secondary structure elements are derived from the crystal structure of DRB7.2M (PDB ID: 8IGD). Identical residues are marked with red highlights, whereas similar residues are marked with yellow highlights, and the consensus residues (> 50%) are annotated below the sequence alignment.
Moreover, the residues involved in the heterodimerization interface in DRB7.2M are identical to those in DRB7.1. As a matter of fact, the residues involved in the dimerization interface, namely, T131, V132, E134, F136, W156, and V161 in DRB7.2M are unchanged in DRB7.1, suggesting that DRB7.1M may interact with DRB4D3 using a similar manner as illustrated for DRB7.2M:DRB4D3 in the manuscript.
Future studies will shed more light on the binding preference of DRB4D3 with DRB7.1 versus DRB7.2. One interesting thing to note is that DRB7.2 is exclusively present in the nucleus, whereas DRB7.1 is observed to localize in the nucleus as well as the cytoplasm. Therefore, spatial restriction may be one of the mechanisms that bring exclusivity to the interaction partner despite having a conserved interaction interface.
Minor comments:
(1) There are no errors for the N, dH, and dS values of the ITC measurements in Table 1. Also, it seems that the measurements are done only once. Values derived from at least triplicates should be presented. This would be helpful to increase confidence in the values derived from ITC, especially for the titration between DRB7.2, DRB4C, and DRB4D3, as the N value there is substantially lower than 1, which does not agree with the other data.
We plan to estimate the errors as proposed by the reviewer in the revised manuscript to ensure that the presented data is of high confidence.
Reviewer #2 (Public review):
Summary:
The manuscript by Paturi and colleagues uses an approach that combines structural biology and biochemistry to probe protein-protein and protein-RNA interactions for two protein factors related to the dsRNA pathway in plants.
Strengths:
A key finding in the research is the direct demonstration of the ability of the single dsRBD (double-strand RNA binding domain) of DRB7.2 to interact simultaneously with dsRNA as well as the C-terminal domain of DRB4. The heterodimerization of DRB7.2 and DRB4 is demonstrated to make a high-affinity complex with dsRNA, and it is proposed that this atypical use of the dsRBD domain to bridge the protein and RNA may contribute to the ability to prevent cleavage that would otherwise occur for dsRNA. The primary results for the interactions are generally well-supported by the data, and the conclusions are taken from the available results without excessive speculation.
Weaknesses:
There is a need for some statistical repeats, as well as a suggested movement of many protein characterization findings in the solution state to support data or to better indicate how these properties could play a role in the final proposed mechanism. There is also the need for certain measurement replicates, such as for the ITC data, which are derived from single measurements and lack sufficient estimates of error.
We plan to restructure the manuscript on the lines proposed by the reviewer in the revised version. Moreover, as mentioned in the response to the comments of Reviewer 1, we suggest estimating the errors to ensure that the presented data is of high confidence in the revised version.
-

-

-