MED26-enriched condensates drive erythropoiesis through modulating transcription pausing
Curation statements for this article:-
Curated by eLife
eLife Assessment
The study is important to show the role of MED26 in red cell formation. Linking transcription pausing with erythropoiesis is a key discovery. The data are solid although there are still spaces to improve. The in vivo data are limited by specificity concerns on their Cre model. Having RNA-seq, using more erythroid markers such as band3 and a4-integrin, and orthogonal validation with iPSC-erythropoiesis model will improve the study.
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Abstract
The Mediator complex regulates various aspects of hematopoietic development, but whether composition of the Mediator complex undergoes dynamic changes for diversifying transcription and functional outputs is unknown. Here, we found that MED26, a subunit in the core Mediator complex, played a distinctive role in facilitating transcription pausing essential for erythroid development. While most Mediator subunits drastically decreased during this process, MED26 remained relatively abundant. Intriguingly, in the early stages, more than half of MED26 occupancy sites did not co-localize with MED1, a representative Mediator subunit, suggesting these subunits exert context-dependent gene regulation. We revealed that MED26-enriched loci were associated with RNA polymerase Ⅱ pausing. MED26 manifested a markedly preferential recruitment of pausing-related factors, leading to an increase in Pol Ⅱ pausing critical for genome-wide transcription repression during erythropoiesis. Moreover, MED26 exhibited pronounced condensate-forming capability, which was necessary for its function in promoting erythropoiesis and recruiting pausing-related factors. Collectively, this study provides mechanistic insights into the functional coordination of distinct Mediator subunits during development and highlights the switch of transcription condensates towards a MED26 enriched form, which modulates transcription pausing to facilitate transcription repression and erythroid development.
Article activity feed
-

eLife Assessment
The study is important to show the role of MED26 in red cell formation. Linking transcription pausing with erythropoiesis is a key discovery. The data are solid although there are still spaces to improve. The in vivo data are limited by specificity concerns on their Cre model. Having RNA-seq, using more erythroid markers such as band3 and a4-integrin, and orthogonal validation with iPSC-erythropoiesis model will improve the study.
-

Reviewer #1 (Public review):
Summary:
In this study from Zhu and colleagues, a clear role for MED26 in mouse and human erythropoiesis is demonstrated that is also mapped to amino acids 88-480 of the human protein. The authors also show the unique expression of MED26 in later-stage erythropoiesis and propose transcriptional pausing and condensate formation mechanisms for MED26's role in promoting erythropoiesis. Despite the author's introductory claim that many questions regarding Pol II pausing in mammalian development remain unanswered, the importance of transcriptional pausing in erythropoiesis has actually already been demonstrated (Martell-Smart, et al. 2023, PMID: 37586368, which the authors notably did not cite in this manuscript). Here, the novelty and strength of this study is MED26 and its unique expression kinetics during …
Reviewer #1 (Public review):
Summary:
In this study from Zhu and colleagues, a clear role for MED26 in mouse and human erythropoiesis is demonstrated that is also mapped to amino acids 88-480 of the human protein. The authors also show the unique expression of MED26 in later-stage erythropoiesis and propose transcriptional pausing and condensate formation mechanisms for MED26's role in promoting erythropoiesis. Despite the author's introductory claim that many questions regarding Pol II pausing in mammalian development remain unanswered, the importance of transcriptional pausing in erythropoiesis has actually already been demonstrated (Martell-Smart, et al. 2023, PMID: 37586368, which the authors notably did not cite in this manuscript). Here, the novelty and strength of this study is MED26 and its unique expression kinetics during erythroid development.
Strengths:
The widespread characterization of kinetics of mediator complex component expression throughout the erythropoietic timeline is excellent and shows the interesting divergence of MED26 expression pattern from many other mediator complex components. The genetic evidence in conditional knockout mice for erythropoiesis requiring MED26 is outstanding. These are completely new models from the investigators and are an impressive amount of work to have both EpoR-driven deletion and inducible deletion. The effect on red cell number is strong in both. The genetic over-expression experiments are also quite impressive, especially the investigators' structure-function mapping in primary cells. Overall the data is quite convincing regarding the genetic requirement for MED26. The authors should be commended for demonstrating this in multiple rigorous ways.
Weaknesses:
(1) The authors state that MED26 was nominated for study based on RNA-seq analysis of a prior published dataset. They do not however display any of that RNA-seq analysis with regards to Mediator complex subunits. While they do a good job showing protein-level analysis during erythropoiesis for several subunits, the RNA-seq analysis would allow them to show the developmental expression dynamics of all subunit members.
(2) The authors use an EpoR Cre for red cell-specific MED26 deletion. However, other studies have now shown that the EpoR Cre can also lead to recombination in the macrophage lineage, which clouds some of the in vivo conclusions for erythroid specificity. That being said, the in vitro erythropoiesis experiments here are convincing that there is a major erythroid-intrinsic effect.
(3) The donor chimerism assessment of mice transplanted with MED26 knockout cells is a bit troubling. First, there are no staining controls shown and the full gating strategy is not shown. Furthermore, the authors use the CD45.1/CD45.2 system to differentiate between donor and recipient cells in erythroblasts. However, CD45 is not expressed from the CD235a+ stage of erythropoiesis onwards, so it is unclear how the authors are detecting essentially zero CD45-negative cells in the erythroblast compartment. This is quite odd and raises questions about the results. That being said, the red cell indices in the mice are the much more convincing data.
(4) The authors make heavy use of defining "erythroid gene" sets and "non-erythroid gene" sets, but it is unclear what those lists of genes actually are. This makes it hard to assess any claims made about erythroid and non-erythroid genes.
(5) Overall the data regarding condensate formation is difficult to interpret and is the weakest part of this paper. It is also unclear how studies of in vitro condensate formation or studies in 293T or K562 cells can truly relate to highly specialized erythroid biology. This does not detract from the major findings regarding genetic requirements of MED26 in erythropoiesis.
(6) For many figures, there are some panels where conclusions are drawn, but no statistical quantification of whether a difference is significant or not.
-

Reviewer #2 (Public review):
Summary:
The manuscript by Zhu et al describes a novel role for MED26, a subunit of the Mediator complex, in erythroid development. The authors have discovered that MED26 promotes transcriptional pausing of RNA Pol II, by recruiting pausing-related factors.
Strengths:
This is a well-executed study. The authors have employed a range of cutting-edge and appropriate techniques to generate their data, including: CUT&Tag to profile chromatin changes and mediator complex distribution; nuclear run-on sequencing (PRO-seq) to study Pol II dynamics; knockout mice to determine the phenotype of MED26 perturbation in vivo; an ex vivo erythroid differentiation system to perform additional, important, biochemical and perturbation experiments; immunoprecipitation mass spectrometry (IP-MS); and the "optoDroplet" assay to …
Reviewer #2 (Public review):
Summary:
The manuscript by Zhu et al describes a novel role for MED26, a subunit of the Mediator complex, in erythroid development. The authors have discovered that MED26 promotes transcriptional pausing of RNA Pol II, by recruiting pausing-related factors.
Strengths:
This is a well-executed study. The authors have employed a range of cutting-edge and appropriate techniques to generate their data, including: CUT&Tag to profile chromatin changes and mediator complex distribution; nuclear run-on sequencing (PRO-seq) to study Pol II dynamics; knockout mice to determine the phenotype of MED26 perturbation in vivo; an ex vivo erythroid differentiation system to perform additional, important, biochemical and perturbation experiments; immunoprecipitation mass spectrometry (IP-MS); and the "optoDroplet" assay to study phase-separation and molecular condensates.
This is a real highlight of the study. The authors have managed to generate a comprehensive picture by employing these multiple techniques. In doing so, they have also managed to provide greater molecular insight into the workings of the MEDIATOR complex, an important multi-protein complex that plays an important role in a range of biological contexts. The insights the authors have uncovered for different subunits in erythropoiesis will very likely have ramifications in many other settings, in both healthy biology and disease contexts.
Weaknesses:
There are almost no discernible weaknesses in the techniques used, nor the interpretation of the data. The IP-MS data was generated in HEK293 cells when it could have been performed in the human CD34+ HSPC system that they employed to generate a number of the other data. This would have been a more natural setting and would have enabled a more like-for-like comparison with the other data.
-

Reviewer #3 (Public review):
Summary:
The authors aim to explore whether other subunits besides MED1 exert specific functions during the process of terminal erythropoiesis with global gene repression, and finally they demonstrated that MED26-enriched condensates drive erythropoiesis through modulating transcription pausing.
Strengths:
Through both in vitro and in vivo models, the authors showed that while MED1 and MED26 co-occupy a plethora of genes important for cell survival and proliferation at the HSPC stage, MED26 preferentially marks erythroid genes and recruits pausing-related factors for cell fate specification. Gradually, MED26 becomes the dominant factor in shaping the composition of transcription condensates and transforms the chromatin towards a repressive yet permissive state, achieving global transcription repression in …
Reviewer #3 (Public review):
Summary:
The authors aim to explore whether other subunits besides MED1 exert specific functions during the process of terminal erythropoiesis with global gene repression, and finally they demonstrated that MED26-enriched condensates drive erythropoiesis through modulating transcription pausing.
Strengths:
Through both in vitro and in vivo models, the authors showed that while MED1 and MED26 co-occupy a plethora of genes important for cell survival and proliferation at the HSPC stage, MED26 preferentially marks erythroid genes and recruits pausing-related factors for cell fate specification. Gradually, MED26 becomes the dominant factor in shaping the composition of transcription condensates and transforms the chromatin towards a repressive yet permissive state, achieving global transcription repression in erythropoiesis.
Weaknesses:
In the in vitro model, the author only used CD34+ cell-derived erythropoiesis as the validation, which is relatively simple, and more in vitro erythropoiesis models need to be used to strengthen the conclusion.
-

Author response:
Public Reviews:
Reviewer #1 (Public review):
Summary:
In this study from Zhu and colleagues, a clear role for MED26 in mouse and human erythropoiesis is demonstrated that is also mapped to amino acids 88-480 of the human protein. The authors also show the unique expression of MED26 in later-stage erythropoiesis and propose transcriptional pausing and condensate formation mechanisms for MED26's role in promoting erythropoiesis. Despite the author's introductory claim that many questions regarding Pol II pausing in mammalian development remain unanswered, the importance of transcriptional pausing in erythropoiesis has actually already been demonstrated (Martell-Smart, et al. 2023, PMID: 37586368, which the authors notably did not cite in this manuscript). Here, the novelty and strength of this study is MED26 and its …
Author response:
Public Reviews:
Reviewer #1 (Public review):
Summary:
In this study from Zhu and colleagues, a clear role for MED26 in mouse and human erythropoiesis is demonstrated that is also mapped to amino acids 88-480 of the human protein. The authors also show the unique expression of MED26 in later-stage erythropoiesis and propose transcriptional pausing and condensate formation mechanisms for MED26's role in promoting erythropoiesis. Despite the author's introductory claim that many questions regarding Pol II pausing in mammalian development remain unanswered, the importance of transcriptional pausing in erythropoiesis has actually already been demonstrated (Martell-Smart, et al. 2023, PMID: 37586368, which the authors notably did not cite in this manuscript). Here, the novelty and strength of this study is MED26 and its unique expression kinetics during erythroid development.
Strengths:
The widespread characterization of kinetics of mediator complex component expression throughout the erythropoietic timeline is excellent and shows the interesting divergence of MED26 expression pattern from many other mediator complex components. The genetic evidence in conditional knockout mice for erythropoiesis requiring MED26 is outstanding. These are completely new models from the investigators and are an impressive amount of work to have both EpoR-driven deletion and inducible deletion. The effect on red cell number is strong in both. The genetic over-expression experiments are also quite impressive, especially the investigators' structure-function mapping in primary cells. Overall the data is quite convincing regarding the genetic requirement for MED26. The authors should be commended for demonstrating this in multiple rigorous ways.
Thank you for your positive feedback.
Weaknesses:
(1) The authors state that MED26 was nominated for study based on RNA-seq analysis of a prior published dataset. They do not however display any of that RNA-seq analysis with regards to Mediator complex subunits. While they do a good job showing protein-level analysis during erythropoiesis for several subunits, the RNA-seq analysis would allow them to show the developmental expression dynamics of all subunit members.
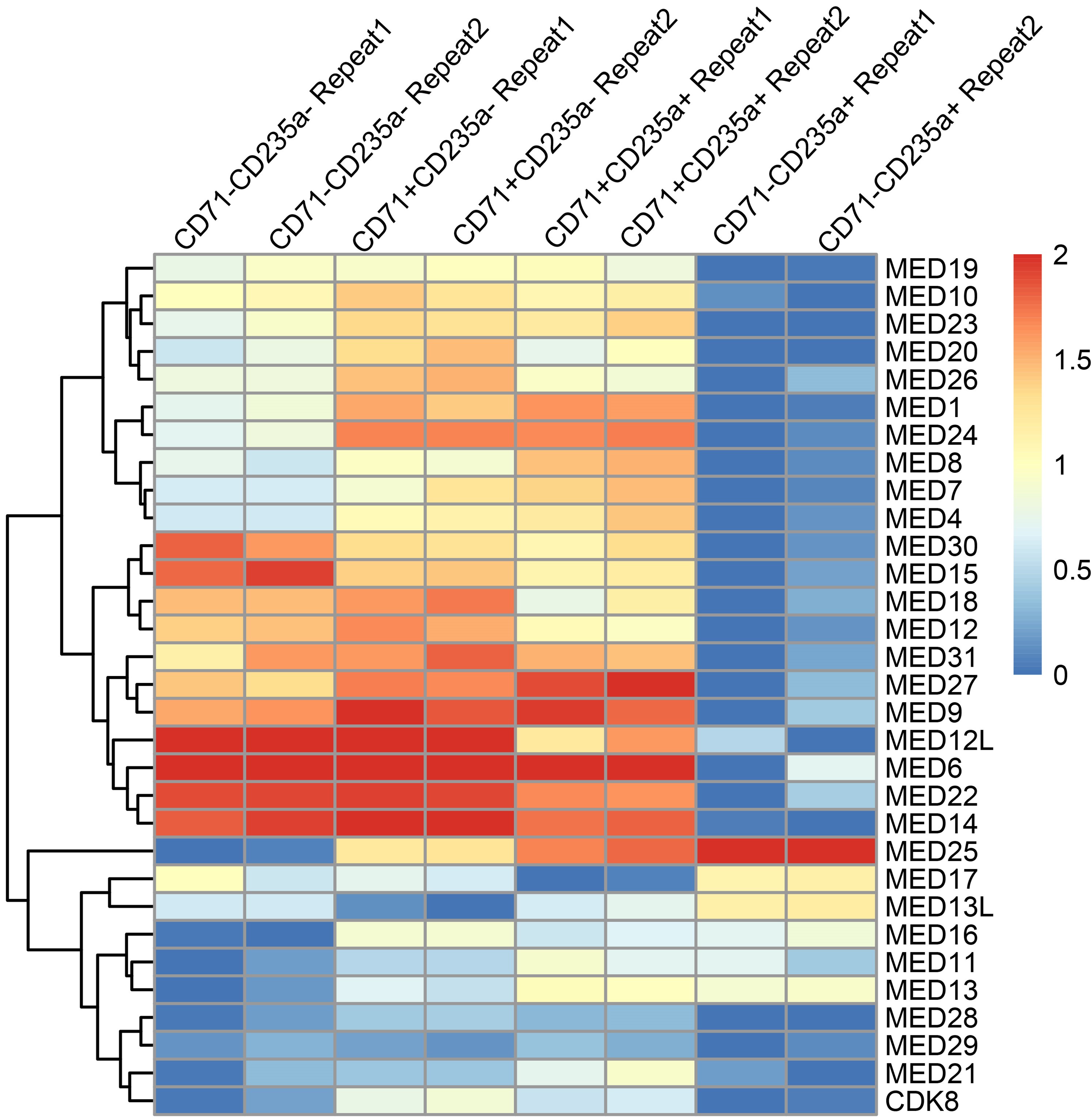
Thank you for this helpful suggestion. While we did not originally nominate MED26 based on RNA-seq analysis, we have analyzed the transcript levels of Mediator complex subunits in our RNA-seq data across different stages of erythroid differentiation (Author response image 1). The results indicate that most Mediator subunits, including MED26, display decreased RNA expression over the course of differentiation, with the exception of MED25, as reported previously (Pope et al., Mol Cell Biol 2013. PMID: 23459945).
Notably, our study is based on initial observations at the protein level, where we found that, unlike most other Mediator subunits that are downregulated during erythropoiesis, MED26 remains relatively abundant. Protein expression levels more directly reflect the combined influences of transcription, translation and degradation processes within cells, and are likely more closely related to biological functions in this context. It is possible that post-transcriptional regulation (such as m6A-mediated improvement of translational efficiency) or post-translational modifications (like escape from ubiquitination) could contribute to the sustained levels of MED26 protein, and this will be an interesting direction for future investigation.
Author response image 1.
Relative RNA expression of Mediator complex subunits during erythropoiesis in human CD34+ erythroid cultures. Different differentiation stages from HSPCs to late erythroblasts were identified using CD71 and CD235a markers, progressing sequentially as CD71-CD235a-, CD71+CD235a-, CD71+CD235a+, and CD71-CD235a+. Expression levels were presented as TPM (transcripts per million).
(2) The authors use an EpoR Cre for red cell-specific MED26 deletion. However, other studies have now shown that the EpoR Cre can also lead to recombination in the macrophage lineage, which clouds some of the in vivo conclusions for erythroid specificity. That being said, the in vitro erythropoiesis experiments here are convincing that there is a major erythroid-intrinsic effect.
Thank you for this insightful comment. We recognize that EpoR-Cre can drive recombination in both erythroid and macrophage lineages (Zhang et al., Blood 2021, PMID: 34098576). However, EpoR-Cre remains the most widely used Cre for studying erythroid lineage effects in the hematopoietic community. Numerous studies have employed EpoR-Cre for erythroid-specific gene knockout models (Pang et al, Mol Cell Biol 2021, PMID: 22566683; Santana-Codina et al., Haematologica 2019, PMID: 30630985; Xu et al., Science 2013, PMID: 21998251.).
While a GYPA (CD235a)-Cre model with erythroid specificity has recently been developed (https://www.sciencedirect.com/science/article/pii/S0006497121029074), it has not yet been officially published. We look forward to utilizing the GYPA-Cre model for future studies. As you noted, our in vivo mouse model and primary human CD34+ erythroid differentiation system both demonstrate that MED26 is essential for erythropoiesis, suggesting that the regulatory effects of MED26 in our study are predominantly erythroid-intrinsic.
(3) Te donor chimerism assessment of mice transplanted with MED26 knockout cells is a bit troubling. First, there are no staining controls shown and the full gating strategy is not shown. Furthermore, the authors use the CD45.1/CD45.2 system to differentiate between donor and recipient cells in erythroblasts. However, CD45 is not expressed from the CD235a+ stage of erythropoiesis onwards, so it is unclear how the authors are detecting essentially zero CD45-negative cells in the erythroblast compartment. This is quite odd and raises questions about the results. That being said, the red cell indices in the mice are the much more convincing data.
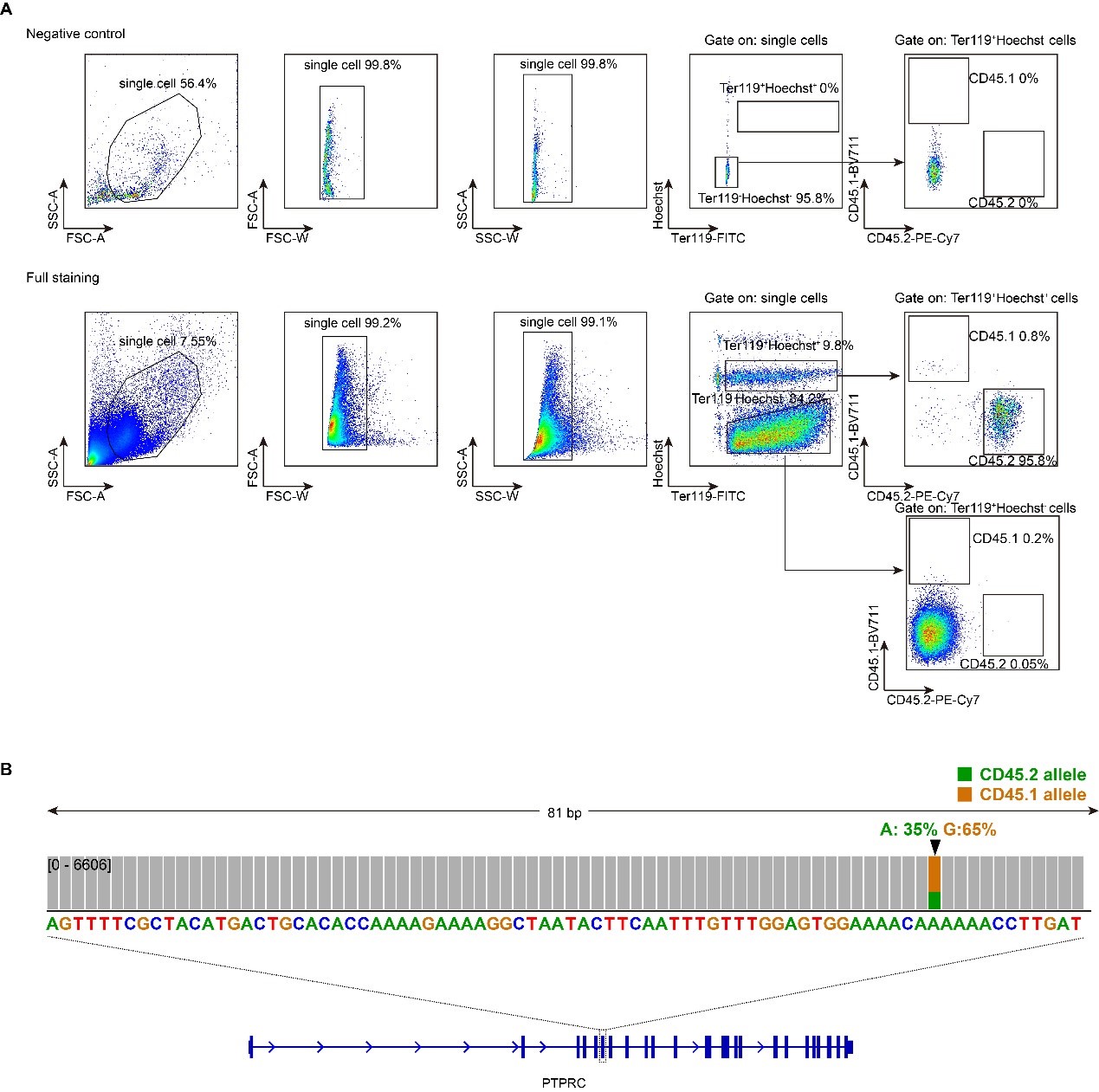
Thank you for your careful and thorough feedback. We have now included negative staining controls (Author response image 2A, top). We agree that CD45 is typically not expressed in erythroid precursors in normal development. Prior studies have characterized BFU-E and CFU-E stages as c-Kit+CD45+Ter119−CD71low and c-Kit+CD45−Ter119−CD71high cells in fetal liver (Katiyar et al, Cells 2023, PMID: 37174702).
However, our observations indicate that erythroid surface markers differ during hematopoiesis reconstitution following bone marrow transplantation. We found that nearly all nucleated erythroid progenitors/precursors (Ter119+Hoechst+) express CD45 after hematopoiesis reconstitution (Author response image 2A, bottom).
To validate our assay, we performed next-generation sequencing by first mixing mouse CD45.1 and CD45.2 total bone marrow cells at a 1:2 ratio. We then isolated nucleated erythroid progenitors/precursors (Ter119+Hoechst+) by FACS and sequenced the CD45 gene locus by targeted sequencing. The resulting CD45 allele distribution matched our initial mixing ratio, confirming the accuracy of our approach (Author response image 2B).
Moreover, a recent study supports that reconstituted erythroid progenitors can indeed be distinguished by CD45 expression following bone marrow transplantation (He et al., Nature Aging 2024, PMID: 38632351. Extended Data Fig. 8).
In conclusion, our data indicate that newly formed erythroid progenitors/precursors post-transplant express CD45, enabling us to identify nucleated erythroid progenitors/precursors by Ter119+Hoechst+ and determine their origin using CD45.1 and CD45.2 markers.
Author response image 2.
Representative flow cytometry gating strategy of erythroid chimerism following mouse bone marrow transplantation. A. Gating strategy used in the erythroid chimerism assay. B. Targeted sequencing result of Ter119+Hoechst+ cells isolated by FACS. The cell sample was pre-mixed with 1/3 CD45.2 and 2/3 CD45.1 bone marrow cells. Ptprc is the gene locus for CD45.
(4) The authors make heavy use of defining "erythroid gene" sets and "non-erythroid gene" sets, but it is unclear what those lists of genes actually are. This makes it hard to assess any claims made about erythroid and non-erythroid genes.
Thank you for this helpful suggestion. We defined "erythroid genes" and "non-erythroid genes" based on RNA-seq data from Ludwig et al. (Cell Reports 2019. PMID: 31189107. Figure 2 and Table S1). Genes downregulated from stages k1 to k5 are classified as “non-erythroid genes,” while genes upregulated from stages k6 to k7 are classified as “erythroid genes.” We will add this description in the revised manuscript.
(5) Overall the data regarding condensate formation is difficult to interpret and is the weakest part of this paper. It is also unclear how studies of in vitro condensate formation or studies in 293T or K562 cells can truly relate to highly specialized erythroid biology. This does not detract from the major findings regarding genetic requirements of MED26 in erythropoiesis.
Thank you for the rigorous feedback. Assessing the condensate properties of MED26 protein in primary CD34+ erythroid cells or mouse models is indeed challenging. As is common in many condensate studies, we used in vitro assays and cellular assays in HEK293T and K562 cells to examine the biophysical properties (Figure S7), condensation formation capacity (Figure 5C and Figure S7C), key phase-separation regions of MED26 protein (Figure S6), and recruitment of pausing factors (Figure 6A-B) in live cells. We then conducted functional assays to demonstrate that the phase-separation region of MED26 can promote erythroid differentiation similarly to the full-length protein in the CD34+ system and K562 cells (Figure 5A). Specifically, overexpressing the MED26 phase-separation domain accelerates erythropoiesis in primary human erythroid culture, while deleting the Intrinsically Disordered Region (IDR) impairs MED26’s ability to form condensates and recruit PAF1 in K562 cells.
In summary, we used HEK293T cells to study the biochemical and biophysical properties of MED26, and the primary CD34+ differentiation system to examine its developmental roles. Our findings support the conclusion that MED26-associated condensate formation promotes erythropoiesis.
(6) For many figures, there are some panels where conclusions are drawn, but no statistical quantification of whether a difference is significant or not.
Thank you for your thorough feedback. We have checked all figures for statistical quantification and added the relevant statistical analysis methods to the corresponding figure legends (Figure 2L and Figure S4C) to clarify the significance of the observed differences. The updated information will be incorporated into the revised manuscript.
Reviewer #2 (Public review):
Summary:
The manuscript by Zhu et al describes a novel role for MED26, a subunit of the Mediator complex, in erythroid development. The authors have discovered that MED26 promotes transcriptional pausing of RNA Pol II, by recruiting pausing-related factors.
Strengths:
This is a well-executed study. The authors have employed a range of cutting-edge and appropriate techniques to generate their data, including: CUT&Tag to profile chromatin changes and mediator complex distribution; nuclear run-on sequencing (PRO-seq) to study Pol II dynamics; knockout mice to determine the phenotype of MED26 perturbation in vivo; an ex vivo erythroid differentiation system to perform additional, important, biochemical and perturbation experiments; immunoprecipitation mass spectrometry (IP-MS); and the "optoDroplet" assay to study phase-separation and molecular condensates.
This is a real highlight of the study. The authors have managed to generate a comprehensive picture by employing these multiple techniques. In doing so, they have also managed to provide greater molecular insight into the workings of the MEDIATOR complex, an important multi-protein complex that plays an important role in a range of biological contexts. The insights the authors have uncovered for different subunits in erythropoiesis will very likely have ramifications in many other settings, in both healthy biology and disease contexts.
Thank you for your thoughtful summary and encouraging feedback.
Weaknesses:
There are almost no discernible weaknesses in the techniques used, nor the interpretation of the data. The IP-MS data was generated in HEK293 cells when it could have been performed in the human CD34+ HSPC system that they employed to generate a number of the other data. This would have been a more natural setting and would have enabled a more like-for-like comparison with the other data.
Thank you for your positive feedback and insightful suggestions. We will perform validation of the immunoprecipitation results in CD34+ derived erythroid cells to further confirm our findings.
Reviewer #3 (Public review):
Summary:
The authors aim to explore whether other subunits besides MED1 exert specific functions during the process of terminal erythropoiesis with global gene repression, and finally they demonstrated that MED26-enriched condensates drive erythropoiesis through modulating transcription pausing.
Strengths:
Through both in vitro and in vivo models, the authors showed that while MED1 and MED26 co-occupy a plethora of genes important for cell survival and proliferation at the HSPC stage, MED26 preferentially marks erythroid genes and recruits pausing-related factors for cell fate specification. Gradually, MED26 becomes the dominant factor in shaping the composition of transcription condensates and transforms the chromatin towards a repressive yet permissive state, achieving global transcription repression in erythropoiesis.
Thank you for your positive summary and feedback.
Weaknesses:
In the in vitro model, the author only used CD34+ cell-derived erythropoiesis as the validation, which is relatively simple, and more in vitro erythropoiesis models need to be used to strengthen the conclusion.
Thank you for your thoughtful suggestions. We have shown that MED26 promotes erythropoiesis using the primary human CD34+ differentiation system (Figure 2 K-M and Figure S4) and have demonstrated its essential role in erythropoiesis through multiple mouse models (Figure 2A-G and Figure S1-3). Together, these in vitro and in vivo results support our conclusion that MED26 regulates erythropoiesis. However, we are open to further validating our findings with additional in vitro erythropoiesis models, such as iPSC or HUDEP erythroid differentiation systems.
-

-

-