Sld3CBD–Cdc45 structural insights into Cdc45 recruitment for CMG complex formation during DNA replication
Curation statements for this article:-
Curated by eLife
eLife Assessment
This valuable paper describes the crystal structure of a complex of the Sld3-Cdc45-binding domain (CBD) with Cdc45, which is essential for the assembly of an active Cdc45-MCM-GINS (CMG) double-hexamer at the replication origin. The structural and biochemical analyses of protein-protein interactions and DNA binding provided solid evidence to support the authors' conclusion. The results shown in the paper are of interest to researchers in DNA replication and genome stability.
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
DNA replication requires recruitment of Cdc45 and GINS into the MCM double hexamer by initiation factors to form an active helicase, the Cdc45–MCM–GINS (CMG) complex, at the replication origins. The initiation factor Sld3 is a central regulator of Cdc45 and GINS recruitment, working with Sld7 together. However, the mechanism through which Sld3 regulates CMG complex formation remains unclear. Here, we present the structure of the Sld3 Cdc45-binding domain in complex with Cdc45 (Sld3CBD–Cdc45), showing detailed interactions and conformational changes required for binding to each other. The mutant analysis indicated that the binding between Sld3CBD and Cdc45 could be broken easily. We also revealed that Sld3CBD, GINS, and MCM bind to different sites on Cdc45 in the Sld3CBD–CMG model, indicating that after recruitment of Cdc45, Sld7–Sld3 could remain in Cdc45–MCM until CMG formation. The consistency between the particle size of Sld7–Sld3–Cdc45 and the distance between Sld3CBDs in the Cdc45–MCM dimer indicated the binding manner of the Cdc45–Sld3–[Sld7] 2 –Sld3–Cdc45 off/on MCM double hexamer. A DNA-binding assay of Sld3 and its complexes with single-stranded ARS1 (autonomously replicating sequence 1) fragments revealed a relationship between the dissociation of Sld7–Sld3 from CMG and the unwound single-stranded DNA. These findings help to further our understanding of the molecular basis of the regulation of CMG complex formation by Sld3.
Article activity feed
-

-

-

-

eLife Assessment
This valuable paper describes the crystal structure of a complex of the Sld3-Cdc45-binding domain (CBD) with Cdc45, which is essential for the assembly of an active Cdc45-MCM-GINS (CMG) double-hexamer at the replication origin. The structural and biochemical analyses of protein-protein interactions and DNA binding provided solid evidence to support the authors' conclusion. The results shown in the paper are of interest to researchers in DNA replication and genome stability.
-

Reviewer #1 (Public review):
Summary:
The crystal structure of the Sld3CBD-Cdc45 complex presented by Li et al. is a significant contribution that enhances our understanding of CMG formation during the rate-limiting step of DNA replication initiation. This structure provides crucial insights into the intermediate steps of CMG formation, and the particle analysis and model predictions compellingly describe the mechanism of Cdc45 loading.
Building upon previously known Sld3 and Cdc45 structures, this study offers new perspectives on how Cdc45 is recruited to MCM DH through the Sld3-Sld7 complex. The most notable finding is the structural rearrangement of Sld3CBD upon Cdc45 binding, particularly the α8-helix conformation, which is essential for Cdc45 interaction and may also be relevant to its metazoan counterpart, Treslin. Additionally, …Reviewer #1 (Public review):
Summary:
The crystal structure of the Sld3CBD-Cdc45 complex presented by Li et al. is a significant contribution that enhances our understanding of CMG formation during the rate-limiting step of DNA replication initiation. This structure provides crucial insights into the intermediate steps of CMG formation, and the particle analysis and model predictions compellingly describe the mechanism of Cdc45 loading.
Building upon previously known Sld3 and Cdc45 structures, this study offers new perspectives on how Cdc45 is recruited to MCM DH through the Sld3-Sld7 complex. The most notable finding is the structural rearrangement of Sld3CBD upon Cdc45 binding, particularly the α8-helix conformation, which is essential for Cdc45 interaction and may also be relevant to its metazoan counterpart, Treslin. Additionally, the conformational shift in the DHHA1 domain of Cdc45 suggests a potential mechanism for its binding to Mcm2NTD.
Furthermore, the ssDNA-binding experiments involving Sld3 further support a broader functional role in the replication process, beyond its established role in recruiting Cdc45. This adds an intriguing new layer to our understanding of Sld3's activity in the yeast. -

Reviewer #2 (Public review):
Summary
The manuscript presents valuable findings, particularly in the crystal structure of the Sld3CBD-Cdc45 interaction and the identification of additional sequences involved in their binding. The modeling of the Sld7-Sld3CBD-CDC45 subcomplex is novel, and the results provide insights into potential conformational changes that occur upon interaction. Although the single-stranded DNA binding data from Sld3 of different species is a minor weakness, the experiments support a model in which the release of Sld3 from the complex may be promoted by its binding to origin single-stranded DNA exposed by the helicase.
-

Reviewer #3 (Public review):
Summary:
The paper by Li et al. describes the crystal structure of a complex of Sld3-Cdc45-binding domain (CBD) with Cdc45 and a model of the dimer of an Sld3-binding protein, Sld7, with two Sld3-CBD-Cdc45 for the tethering. In addition, the authors showed the genetic analysis of the amino acid substitution of residues of Sld3 in the interface with Cdc45 and biochemical analysis of the protein interaction between Sld3 and Cdc45 as well as DNA binding activity of Sld3 to the single-strand DNAs of the ARS sequence.
-

Author response:
The following is the authors’ response to the previous reviews
Reviewer #1 (Public review):
Summary:
The crystal structure of the Sld3CBD-Cdc45 complex presented by Li et al. is a significant contribution that enhances our understanding of CMG formation during the rate-limiting step of DNA replication initiation. This structure provides crucial insights into the intermediate steps of CMG formation, and the particle analysis and model predictions compellingly describe the mechanism of Cdc45 loading. Building upon previously known Sld3 and Cdc45 structures, this study offers new perspectives on how Cdc45 is recruited to MCM DH through the Sld3-Sld7 complex. The most notable finding is the structural rearrangement of Sld3CBD upon Cdc45 binding, particularly the α8-helix conformation, which is essential for Cdc45 …
Author response:
The following is the authors’ response to the previous reviews
Reviewer #1 (Public review):
Summary:
The crystal structure of the Sld3CBD-Cdc45 complex presented by Li et al. is a significant contribution that enhances our understanding of CMG formation during the rate-limiting step of DNA replication initiation. This structure provides crucial insights into the intermediate steps of CMG formation, and the particle analysis and model predictions compellingly describe the mechanism of Cdc45 loading. Building upon previously known Sld3 and Cdc45 structures, this study offers new perspectives on how Cdc45 is recruited to MCM DH through the Sld3-Sld7 complex. The most notable finding is the structural rearrangement of Sld3CBD upon Cdc45 binding, particularly the α8-helix conformation, which is essential for Cdc45 interaction and may also be relevant to its metazoan counterpart, Treslin. Additionally, the conformational shift in the DHHA1 domain of Cdc45 suggests a potential mechanism for its binding to Mcm2NTD. Furthermore, Sld3's ssDNA-binding experiments provide evidence of its novel functions in the DNA replication process in yeast, expanding our understanding of its role beyond Cdc45 recruitment.
Strengths:
The manuscript is generally well-written, with a precise structural analysis and a solid methodological section that will significantly advance future studies in the field. The predictions based on structural alignments are intriguing and provide a new direction for exploring CMG formation, potentially shaping the future of DNA replication research. This research also opens up several new opportunities to utilize structural biology to unravel the molecular details of the model presented in the paper.
Weaknesses:
The main weakness of the manuscript lies in the lack of detailed structural validation for the proposed Sld3-Sld7-Cdc45 model, and its CMG bound models, which could be done in the future using advanced structural biology techniques such as single particle cryo-electron microscopy. It would also be interesting to explore how Sld7 interacts with the MCM helicase, and this would help to build a detailed long-flexible model of Sld3-Sld7-Cdc45 binding to MCM DH and to show where Sld7 will lie on the structure. This will help us to understand how Sld7 functions in the complex. Also, future experiments would be needed to understand the molecular details of how Sld3 and Sld7 release from CMG is associated with ssARS1 binding.
The proposals based on this study provide new knowledge of the CMG formation process. We agree that our Sld3-Sld7-Cdc45 model will be further confirmed by cryo-EM. We improved our ssARS1-binding assay and quantified data (See the response to Recommendations for the authors of #3 review).
Reviewer #2 (Public review):
Summary
The manuscript presents valuable findings, particularly in the crystal structure of the Sld3CBD-Cdc45 interaction and the identification of additional sequences involved in their binding. The modeling of the Sld7-Sld3CBD-CDC45 subcomplex is novel, and the results provide insights into potential conformational changes that occur upon interaction. Although the single-stranded DNA binding data from Sld3 of different species is a minor weakness, the experiments support a model in which the release of Sld3 from the complex may be promoted by its binding to origin single-stranded DNA exposed by the helicase.
Strengths
The Sld3CBD-Cdc45 structure is a novel contribution, revealing critical residues involved in the interaction.
The model structures generated from the crystal data are well presented and provide valuable insights into the interaction sequences between Sld3 and Cdc45.
The experiments testing the requirements for interaction sequences are thorough and conducted well, with clear figures supporting the conclusions.
The conformational changes observed in Sld3 and Cdc45 upon binding are interesting and enhance our understanding of the interaction.
The modeling of the Sld7-Sld3CBD-CDC45 subcomplex is a new and valuable addition to the field.
The proposed model of Sld3 release from the complex through binding to single stranded DNA at the origin is intriguing.
Weaknesses
The section on the binding of Sld3 complexes to origin single-stranded DNA is somewhat weakened by the use of Sld3 proteins from different species. The comparisons between Sld3-CBD, Sld3CBD-Cdc45, and Sld7-Sld3CBD-Cdc45 involve complexes from different species, limiting the comparisons' value.
Although the study reveals that Sld3 binds to different residues of Cdc45 than those previously shown to bind Mcm or GINS, the data in the paper do not shed any additional light on how GINS and Sld3 binding to Cdc45 or Mcms. would affect each other. Other previous research has suggested that the binding of GINS and Sld3 to Mcm or Cdc45 may be mutually exclusive. The authors acknowledge that a structural investigation of Sld3, Sld7, Cdc45, and MCM during the stage of GINS recruitment will be a significant goal for future research.
We agree that it is better to use all samples from a source; however, due to limitations in protein expression, we used Sld7-Sld3CBD-Cdc45 from a different source. The two sources used in this study belong to the same family, and the proteins Sld7, Sld3 and Cdc45 share sequence conservation with similar structures predicted by Alphafold3 (RMSD = 0.356, 1.392, and 0.891 for Ca atoms of Sld7CTD, Sld7NTD-Sld3NTD, and Sld3CBD-Cdc45). Such similarity in source and proteins allows us to do the comparison. We also mentioned that a cryo-EM study of Sld3-Sld7-Cdc45-MCM and Sld3-Sld7-CMG structures will be a significant goal for future research in our manuscript.
Reviewer #3 (Public review):
Summary:
The paper by Li et al. describes the crystal structure of a complex of Sld3-Cdc45-binding domain (CBD) with Cdc45 and a model of the dimer of an Sld3-binding protein, Sld7, with two Sld3-CBD-Cdc45 for the tethering. In addition, the authors showed the genetic analysis of the amino acid substitution of residues of Sld3 in the interface with Cdc45 and biochemical analysis of the protein interaction between Sld3 and Cdc45 as well as DNA binding activity of Sld3 to the single-strand DNAs of the ARS sequence.
Strengths:
The authors provided a nice model of an intermediate step in the assembly of an active Cdc45-MCM-GINS (CMG) double hexamers at the replication origin, which is mediated by the Sld3-Sld7 complex. The dimer of the Sld3-Sld7 complexes tethers two MCM hexamers together for the recruitment of GINS-Pol epsilon on the replication origin.
Weaknesses:
The biochemical analysis should be carefully evaluated with more quantitative ways to strengthen the authors' conclusion even in the revised version.
In this revision, we improved our ssARS1-binding assay in more quantitative ways (See the response to Recommendations for the authors).
Reviewer #1 (Recommendations for the authors):
I thank the authors for all their replies to my previous questions and for doing all the necessary corrections. I am satisfied with most of their replies, however, upon second reading I have a few more suggestions which could help to improve the manuscript further and make an impact in the field. My comments are listed below.
(1) In general, the manuscript is well structured, but I feel that it requires professional English correction. In many places it was difficult to understand the sentences and I had to read it several times to understand it. Also, very long sentences should be avoided. The flow should be easy to read and understand, and that is why I feel it requires professional English correction.
Following the comment, we checked English carefully and shortened the very long sentences.
(2) Page 5, line 103, please include molecule after the word complex to make it like- "Only one complex molecule exists within an asymmetric unit."
We revised this sentence (P5/L103).
(3) Line 113- more than the N-terminal half of the protruding long helix α7 113 was disordered in the Sld3CBD-Cdc45 complex. This sentence is not clear. What does it mean more than the N-terminal half? Please rewrite it.
We revised this sentence to give the corresponding residue number “(D219–H231)” (P5/L114).
(4) Page 5, result 2- Conformation changes in Sld3CBD and Cdc45 for binding each other, this section may require a little restructuring. Line 130-131- "Therefore, the helix α8CTP seems to be an intrinsically disordered segment when Sld3 alone but 130 folds into a helix coupled to the binding partner Cdc45 in the Sld3CBD-Cdc45 complex." This statement is the crux of the structural finding and therefore, I feel it should move after the first sentence.
Thank you for your comments. We rewrote this part (P5/L128-131).
(5) Line 121-122: Compared to the isolated form (PDBIDs: 5DGO 121 for huCdc45 [31] and 6CC2 for EhCdc45 [33]) and the CMG form (PDBID: 3JC6. Write it in the same format. Make 6CC2 in bracket like other PDB IDs. Restructure this sentence.
We revised this sentence (P5/122-123).
(6) Line 127-129: This sentence is also not very clear.
We revised this sentence together with above No (4). (P5/L128-131)
(7) In my question 4- "Can authors add a supplementary figure showing the probability of disordernes..."., I meant to use a disorder prediction tool like IUPred for the protein sequences and show that α8 is predicted to be a disordered upon sequence analysis. This will help to show the inherent property of α8 helix, and it could add up to the understanding that a disordered region is being structured in the complex structure.
The structures showed that α8CTP is stabilized by binding with Cdc45, but disordered in Sld3CBD alone, indicating that this part is flexible, like an intrinsically disordered segment. We have deposited the structure to PDB, so predictions like IUPred cannot show meaningful information.
(8) Question 9 regarding Supplementary Figure 8- Please include your statement in the figure legend - "WT Sld3CBD was prepared in a complex with Cdc45, while the mutants of Sld3CBD existed alone, we calculated the elements of secondary structure from the crystal structure of Sld3CBD-Cdc45. The concentration of samples was controlled to the same level for CD measurement."
Following the comment, we optimized the figure legend of Supplementary Figure 8.
(9) Question 13- I understand that negative staining and SEC-SAXS experiments could be very tricky for such protein complexes, which have very long loops and are flexible. Did authors try a GraFix cross-linking before doing the negative staining TEM? If it is not being tried, then it might be a good idea to try it and it may help to get much cleaner particles and easier class averaging. Although I completely understand the technical challenges the authors describe and I agree with them, I still feel that one good experiment that shows this dimer model would be very helpful to strengthen the claim. I am concerned because if people start using a similar DLS experiment to calculate intermolecular distances, citing your paper, in many cases it might be a wrong interpretation. In case the negative staining still does not work, at least discuss your technical challenges in the discussion section and mention that SEC-SAXS showed a similar length of the complex and show the Guinier plot and Porod plots in the supplementary data.
We believe that DLS is one of the methods for analyzing the single particle size. Of course, the confirmation by multiple methods will give compelling evidence. Following the comment, we added SEC-SAXS data in the [Results] (P7/L194-196) (Cdc45 recruitment to MCM DH by Sld3 with partner Sld7) and Supplementary Figure 11. The Sld7-Sld3-Cdc45 forms a flexible, long shape. Each binding domain is rigid but linked by the long loops. The flexibility problems are caused by the long loop linkers, but not by binding. So, we did not try to use the cross-linking method for analysis experiments.
(10) Page 8, line 221- litter sequence specificity: Correct the word "litter" with little. Also, the word shaped is written as sharped at a few places in the manuscript. Please correct it.
We apologize for making such mistakes. We have modified these words.
(11) Page 9, line 237-238: Would it be possible to add a lane showing Sld7 binding to the ssDNA in figure 4. I recommend showing this to understand the ssDNA binding affinity of Sld7 by itself and it will also help us to compare when it is in complex with Sld3.
Considering that Sld7 on CMG is always a complex with Sld3, the ssDNA binding affinity should use the Sld3-Sld7 complex. Additionally, we attempted to overexpress Sld7, but could not obtain the target protein.
Reviewer #2 (Recommendations for the authors):
Thank you for the improved manuscript. The following sentence is unclear: "Cdc45 binds tighter to long ssDNA (>60 bases) with a litter sequence specificity".
We apologize for making such a mistake. We modified “litter” to “little”.
I found it challenging to understand which species were used while reading the results section and figure legends. I recommend that the authors revise the text in both the results and figure legends to clearly indicate when proteins from different species are being compared. Additionally, it would be valuable to explicitly acknowledge this limitation in the text.
Following the comment, we added a description for using different species in results (P8/L224-225) and figure legends (Supplementary Figure 14). We added more information in the Methods to explain why we used two species for preparing proteins.
Reviewer #3 (Recommendations for the authors):
Major points:
(1) The current title is not appropriate for the general readers. At least, DNA replication or DNA replication initiation should be added and abbreviations such as CBD should be avoided.
Following the comment, we added “DNA replication” into the title. Regarding “CBD”, since the full name of “Cdc45 binding domain” is too long, we continue to use Sld3CBD.
(2) As in my previous review, I asked for quantification of the EMSA assay shown in Figure 4 and Supplemental Figures 13 and 14. Since some signals of the bands are very weak, it is hard to conclude something. Given different protein concentrations used in the experiment, the authors should provide any kinds of value. For example, Sld3CBD-CDC45 shows weaker DNA binding than Sld3CBD alone (line 231). Is this true (or reproducible)? It is hard to conclude without any quantification.
We have repeated the EMSA assay four or more times with different rods of overexpression, purification and DNA synthesis, indicating that the EMSA assay is reproducible. In this revision, we changed the DNA stain and adjusted the ratio between the protein and ssDNA with increasing concentrations. The smeared bands of ssDNA with Sld7–Sld3ΔC–Cdc45 or Sld7–Sld3ΔC exhibit enhanced discernibility, and the ssDNA bands are intense enough for grayscale calculations (Figure 4 in the second revised version). We used a series of t-tests to confirm a significantly ssDNA residual level between Sld3CBD–Cdc45 to Sld3CBD, Sld7–Sld3ΔC–Cdc45, and Sld7–Sld3ΔCS (t-test, ****: P<0.0001). We also carefully controlled the sample amount in the EMAS assay and described it in the [Methods].
Moreover, in this EMSA assay (in Figure 4), the authors suggest that the disappearance of ssDNA bands corresponds with the binding of the protein to the DNA. However, it is also possible that the DNA is degraded. It is very important to show the band of protein-DNA complexes on the gel (a whole gel, not the parts of the gel shown in Figure). Why did the authors use this "insensitive" assay using SyberGreen, not radio-labelled ssDNA?
In this revision, we added a negative control of no ssDNA-binding by using ssARS1-3_3 for all protein samples (Sld3CBD, Sld3CBD–Cdc45, Sld7–Sld3ΔC–Cdc45 and Sld7–Sld3ΔC), which were the same rod of expression and purification for bound to ssARS1s (ssARS1-2 and ssARS1-5) (Figure 4), showing that the disappearance of ssDNA bands is caused by binding to proteins, not degradation. Moreover, this time, by changing the DNA stain and increasing the concentration of the samples, the smeared ssDNA bands exhibit enhanced discernibility in the high molecular weight regions when mixed with Sld7–Sld3ΔC–Cdc45 or Sld7–Sld3ΔC, whereas no bands appeared in the NC (ssARS1-3_1). The positions of smeared ssDNA bonds correspond to those of protein in the protein-stain pages, indicating that ssARS1 were complexed with proteins. Following the comment, we show all bands on the gel in Figure 4 and Supplementary Figure 14. Compared to Sld7–Sld3ΔC–Cdc45 or Sld7–Sld3ΔC, Sld3CBD and ssDNA bonds could not be observed because the pI value of Sld3CBD, which affects the entry of the samples into the gel.
We agree that using radio-labelled ssDNA can obtain a sensitive binding assay. However, current laboratory constraints did not allow us to use radio-labelled ssDNA. Furthermore, considering the characteristics of our target proteins, Sld3CBD, Sld3CBD–Cdc45, Sld7–Sld3ΔC–Cdc45, and Sld7–Sld3ΔC, we planned to perform the binding assay in a more natural state without any modifications, labelling or linkers. Additionally, we have attempted to use ITC experiments but failed in the measurements. Presumably, the conformational flexibility of Sld7-Sld3-Cdc45 and Sld7-Sld3 caused a thermodynamic anomaly.
Minor points:
(1) Line 215, 80b: This should be "80 nucleotides(nt)". Throughout the text, nucleotides is better than base to show the length of ssDNAs.
Thank you for your comments. We modified these words throughout the text.
-

eLife Assessment
This valuable paper describes the crystal structure of a complex of Sld3-Cdc45-binding domain (CBD) with Cdc45, which is essential for the assembly of an active Cdc45-MCM-GINS (CMG) double hexamers at the replication origin. Although the results shown in the paper are of interest to researchers in DNA replication and genome stability, the biochemical analysis of protein-protein interaction and DNA binding is incomplete, and the paper needs additional data.
-

Reviewer #1 (Public review):
Summary:
The crystal structure of the Sld3CBD-Cdc45 complex presented by Li et al. is a significant contribution that enhances our understanding of CMG formation during the rate-limiting step of DNA replication initiation. This structure provides crucial insights into the intermediate steps of CMG formation, and the particle analysis and model predictions compellingly describe the mechanism of Cdc45 loading. Building upon previously known Sld3 and Cdc45 structures, this study offers new perspectives on how Cdc45 is recruited to MCM DH through the Sld3-Sld7 complex. The most notable finding is the structural rearrangement of Sld3CBD upon Cdc45 binding, particularly the α8-helix conformation, which is essential for Cdc45 interaction and may also be relevant to its metazoan counterpart, Treslin. Additionally, …
Reviewer #1 (Public review):
Summary:
The crystal structure of the Sld3CBD-Cdc45 complex presented by Li et al. is a significant contribution that enhances our understanding of CMG formation during the rate-limiting step of DNA replication initiation. This structure provides crucial insights into the intermediate steps of CMG formation, and the particle analysis and model predictions compellingly describe the mechanism of Cdc45 loading. Building upon previously known Sld3 and Cdc45 structures, this study offers new perspectives on how Cdc45 is recruited to MCM DH through the Sld3-Sld7 complex. The most notable finding is the structural rearrangement of Sld3CBD upon Cdc45 binding, particularly the α8-helix conformation, which is essential for Cdc45 interaction and may also be relevant to its metazoan counterpart, Treslin. Additionally, the conformational shift in the DHHA1 domain of Cdc45 suggests a potential mechanism for its binding to Mcm2NTD. Furthermore, Sld3's ssDNA-binding experiments provide evidence of its novel functions in the DNA replication process in yeast, expanding our understanding of its role beyond Cdc45 recruitment.
Strengths:
The manuscript is generally well-written, with a precise structural analysis and a solid methodological section that will significantly advance future studies in the field. The predictions based on structural alignments are intriguing and provide a new direction for exploring CMG formation, potentially shaping the future of DNA replication research. This research also opens up several new opportunities to utilize structural biology to unravel the molecular details of the model presented in the paper.
Weaknesses:
The main weakness of the manuscript lies in the lack of detailed structural validation for the proposed Sld3-Sld7-Cdc45 model, and its CMG bound models, which could be done in the future using advanced structural biology techniques such as single particle cryo-electron microscopy. It would also be interesting to explore how Sld7 interacts with the MCM helicase, and this would help to build a detailed long-flexible model of Sld3-Sld7-Cdc45 binding to MCM DH and to show where Sld7 will lie on the structure. This will help us to understand how Sld7 functions in the complex. Also, future experiments would be needed to understand the molecular details of how Sld3 and Sld7 release from CMG is associated with ssARS1 binding.
-

Reviewer #2 (Public review):
Summary
The manuscript presents valuable findings, particularly in the crystal structure of the Sld3CBD-Cdc45 interaction and the identification of additional sequences involved in their binding. The modeling of the Sld7-Sld3CBD-CDC45 subcomplex is novel, and the results provide insights into potential conformational changes that occur upon interaction. Although the single-stranded DNA binding data from Sld3 of different species is a minor weakness, the experiments support a model in which the release of Sld3 from the complex may be promoted by its binding to origin single-stranded DNA exposed by the helicase.
Strengths
• The Sld3CBD-Cdc45 structure is a novel contribution, revealing critical residues involved in the interaction.
• The model structures generated from the crystal data are well presented and …Reviewer #2 (Public review):
Summary
The manuscript presents valuable findings, particularly in the crystal structure of the Sld3CBD-Cdc45 interaction and the identification of additional sequences involved in their binding. The modeling of the Sld7-Sld3CBD-CDC45 subcomplex is novel, and the results provide insights into potential conformational changes that occur upon interaction. Although the single-stranded DNA binding data from Sld3 of different species is a minor weakness, the experiments support a model in which the release of Sld3 from the complex may be promoted by its binding to origin single-stranded DNA exposed by the helicase.
Strengths
• The Sld3CBD-Cdc45 structure is a novel contribution, revealing critical residues involved in the interaction.
• The model structures generated from the crystal data are well presented and provide valuable insights into the interaction sequences between Sld3 and Cdc45.
• The experiments testing the requirements for interaction sequences are thorough and conducted well, with clear figures supporting the conclusions.
• The conformational changes observed in Sld3 and Cdc45 upon binding are interesting and enhance our understanding of the interaction.
• The modeling of the Sld7-Sld3CBD-CDC45 subcomplex is a new and valuable addition to the field.
• The proposed model of Sld3 release from the complex through binding to single stranded DNA at the origin is intriguing.Weaknesses
• The section on the binding of Sld3 complexes to origin single-stranded DNA is somewhat weakened by the use of Sld3 proteins from different species. The comparisons between Sld3-CBD, Sld3CBD-Cdc45, and Sld7-Sld3CBD-Cdc45 involve complexes from different species, limiting the comparisons' value.
• Although the study reveals that Sld3 binds to different residues of Cdc45 than those previously shown to bind Mcm or GINS, the data in the paper do not shed any additional light on how GINS and Sld3 binding to Cdc45 or Mcms. would affect each other. Other previous research has suggested that the binding of GINS and Sld3 to Mcm or Cdc45 may be mutually exclusive. The authors acknowledge that a structural investigation of Sld3, Sld7, Cdc45, and MCM during the stage of GINS recruitment will be a significant goal for future research. -

Reviewer #3 (Public review):
Summary:
The paper by Li et al. describes the crystal structure of a complex of Sld3-Cdc45-binding domain (CBD) with Cdc45 and a model of the dimer of an Sld3-binding protein, Sld7, with two Sld3-CBD-Cdc45 for the tethering. In addition, the authors showed the genetic analysis of the amino acid substitution of residues of Sld3 in the interface with Cdc45 and biochemical analysis of the protein interaction between Sld3 and Cdc45 as well as DNA binding activity of Sld3 to the single-strand DNAs of the ARS sequence.
Strengths:
The authors provided a nice model of an intermediate step in the assembly of an active Cdc45-MCM-GINS (CMG) double hexamers at the replication origin, which is mediated by the Sld3-Sld7 complex. The dimer of the Sld3-Sld7 complexes tethers two MCM hexamers together for the recruitment of …
Reviewer #3 (Public review):
Summary:
The paper by Li et al. describes the crystal structure of a complex of Sld3-Cdc45-binding domain (CBD) with Cdc45 and a model of the dimer of an Sld3-binding protein, Sld7, with two Sld3-CBD-Cdc45 for the tethering. In addition, the authors showed the genetic analysis of the amino acid substitution of residues of Sld3 in the interface with Cdc45 and biochemical analysis of the protein interaction between Sld3 and Cdc45 as well as DNA binding activity of Sld3 to the single-strand DNAs of the ARS sequence.
Strengths:
The authors provided a nice model of an intermediate step in the assembly of an active Cdc45-MCM-GINS (CMG) double hexamers at the replication origin, which is mediated by the Sld3-Sld7 complex. The dimer of the Sld3-Sld7 complexes tethers two MCM hexamers together for the recruitment of GINS-Pol epsilon on the replication origin.
Weaknesses:
The biochemical analysis should be carefully evaluated with more quantitative ways to strengthen the authors' conclusion even in the revised version.
-

Author response:
The following is the authors’ response to the original reviews
Public Reviews:
Reviewer #1 (Public review):
Summary:
The crystal structure of the Sld3CBD-Cdc45 complex presented by Li et al. is a novel contribution that significantly advances our understanding of CMG formation during the rate-limiting step of DNA replication initiation. This structure provides insights into the intermediate steps of CMG formation. The study builds upon previously known structures of Sld3 and Cdc45 and offers new perspectives into how Cdc45 is loaded onto MCM DH through Sld3-Sld7. The most notable finding is the structural difference in Sld3CBD when bound to Cdc45, particularly the arrangement of the α8-helix, which is essential for Cdc45 binding and may also pertain to its metazoan counterpart, Treslin. Additionally, the conformational …
Author response:
The following is the authors’ response to the original reviews
Public Reviews:
Reviewer #1 (Public review):
Summary:
The crystal structure of the Sld3CBD-Cdc45 complex presented by Li et al. is a novel contribution that significantly advances our understanding of CMG formation during the rate-limiting step of DNA replication initiation. This structure provides insights into the intermediate steps of CMG formation. The study builds upon previously known structures of Sld3 and Cdc45 and offers new perspectives into how Cdc45 is loaded onto MCM DH through Sld3-Sld7. The most notable finding is the structural difference in Sld3CBD when bound to Cdc45, particularly the arrangement of the α8-helix, which is essential for Cdc45 binding and may also pertain to its metazoan counterpart, Treslin. Additionally, the conformational shift in the DHHA1 domain of Cdc45 suggests a possible mechanism for its binding to MCM2NTD.
Strengths:
The manuscript is generally well-written, with a precise structural analysis and a solid methodological section that will significantly advance future studies in the field. The predictions based on structural alignments are intriguing and provide a new direction for exploring CMG formation, potentially shaping the future of DNA replication research.
Weaknesses:
The main weakness of the manuscript lies in the lack of experimental validation for the proposed Sld3-Sld7-Cdc45 model. Specifically, the claim that Sld3 binding to Cdc45-MCM does not inhibit GINS binding, a finding that contradicts previous research, is not sufficiently substantiated with experimental evidence. To strengthen their model, the authors must provide additional experimental data to support this mechanism. Also, the authors have not compared the recently published Cryo-EM structures of the metazoan CMG helicases with their predicted models to see if Sld3/Treslin does not cause any clash with the GINS when bound to the CMG. Still, the work holds great potential in its current form but requires further experiments to confirm the authors' conclusions.
We appreciate the reviewers’ careful reading and the comments.
Our structural analysis of Sld3CBD-Cdc45 showed the detailed interaction map between Sld3CBD and Cdc45 at 2.6 Å resolution. The Sld3, MCM and GINS binding sites of Cdc45 completely differed, suggesting that the Sld3CBD, Cdc45 and GINS could bind to MCM together. The SCMG-DNA model confirmed such a binding manner, although our study does not show how this binding manner affects the GINS loading by other initiation factors (Dpb11, Sld2, et. al). Regarding the previous studies, competition of Sld3 and GINS for binding to Cdc45 or Cdc45-MCM (Bruck et. al), which may be caused by the conformation change of Cdc45 DHHA1 between Sld3CBD-Cdc45 and CMG. We modified our manuscript and discussed (P7/L168-173, and P10/L282-286). Following the comment, we checked the recently published Cryo-EM structure (PDBID:8Q6O) with their predicted models of the metazoan CMG helicases (P7/L198-P8/L202) and added the Cdc45 mutation experiments to confirm our conclusion ([Recommendations for the authors] Q18).
Reviewer #2 (Public review):
Summary
The manuscript presents valuable findings, particularly in the crystal structure of the Sld3CBD-Cdc45 interaction and the identification of additional sequences involved in their binding. The modeling of the Sld7-Sld3CBD-CDC45 subcomplex is novel, and the results provide insights into potential conformational changes that occur upon interaction. However, the work remains incomplete as several main claims are only partially supported by experimental data, particularly the proposed model for Sld3 interaction with GINS on the CMG. Additionally, the single-stranded DNA binding data from different species do not convincingly advance the manuscript's central arguments.
Strengths
(1) The Sld3CBD-Cdc45 structure is a novel contribution, revealing critical residues involved in the interaction.
(2) The model structures generated from the crystal data are well presented and provide valuable insights into the interaction sequences between Sld3 and Cdc45.
(3) The experiments testing the requirements for interaction sequences are thorough and conducted well, with clear figures supporting the conclusions.
(4) The conformational changes observed in Sld3 and Cdc45 upon binding are interesting and enhance our understanding of the interaction.
(5) The modeling of the Sld7-Sld3CBD-CDC45 subcomplex is a new and valuable addition to the field.
Weaknesses
(1) The proposed model for Sld3 interacting with GINS on the CMG needs more experimental validation and conflicts with published findings. These discrepancies need more detailed discussion and exploration.
Our structural analysis experiment of Sld3CBD-Cdc45 showed the detailed interaction information between Sld3CBD and Cdc45 at 2.6 Å resolution. The Sld3CBD-binding site of Cdc45 is completely different from that of GINS and MCM binding to Cdc45, suggesting that the Sld3CBD, Cdc45, and GINS could bind to MCM together. The SCMG-DNA model confirmed such a binding manner. Following the comment, we added a Cdc45 mutant analysis, disrupting the binding to MCM and GINS but not affecting the Sld3CBD binding (Supplementary Figure 9). Our model is consistent with the GINS-loading requirement (the phosphorylation of Sld3 on Cdc45-MCM) and has no discrepancies with the stepwise loading fashion (Please see the responses to [Recommendations for the authors] Reviewer#1-Q14-15]). Regarding the previous studies, competition of Sld3 and GINS for binding to Cdc45 or Cdc45-MCM (Bruck et. al), by in vitro binding experiments, please see the responses to [Recommendations for the authors] Q6.
(2) The section on the binding of Sld3 complexes to origin single-stranded DNA needs significant improvement. The comparisons between Sld3-CBD, Sld3CBD-Cdc45, and Sld7-Sld3CBD-Cdc45 involve complexes from different species, limiting the comparisons' value.
As suggested, we tried to improve the ssDNA-binding section (Please see the responses to [Recommendations for the authors]: Q4 and Q5). We used Sld7-Sld3CBD-Cdc45 from different sources due to limitations in protein expression. These two sources belong to the same family and the proteins Sld7, Sld3 and Cdc45 have sequence conservation with similar structures predicted by the alphafold3 (RMSD = 0.356, 1.392, and 0.891 for Ca atoms of Sld7CTD, Sld7NTD-Sld3NTD, and Sld3CBD-Cdc45). Such similarity in source and protein lever allows us to do the comparison.
(3) The authors' model proposing the release of Sld3 from CMG based on its binding to single-stranded DNA is unclear and needs more elaboration.
Considering that ssDNA (ssARS1) is produced by CMG, the ssDNA-binding of Sld3 should happen after forming an active CMG. Therefore, the results of ssDNA binding experiments implied that the Sld3 release could be with the binding to ssDNA produced by CMG. We tried to present more elaborations in the revised version. (Please see the responses to [Recommendations for the authors] Q4, Q5).
Reviewer #3 (Public review):
Summary:
The paper by Li et al. describes the crystal structure of a complex of Sld3-Cdc45-binding domain (CBD) with Cdc45 and a model of the dimer of an Sld3-binding protein, Sld7, with two Sld3-CBD-Cdc45 for the tethering. In addition, the authors showed the genetic analysis of the amino acid substitution of residues of Sld3 in the interface with Cdc45 and biochemical analysis of the protein interaction between Sld3 and Cdc45 as well as DNA binding activity of Sld3 to the single-strand DNAs of the ARS sequence.
Strengths:
The authors provided a nice model of an intermediate step in the assembly of an active Cdc45-MCM-GINS (CMG) double hexamers at the replication origin, which is mediated by the Sld3-Sld7 complex. The dimer of the Sld3-Sld7 complexes tethers two MCM hexamers together for the recruitment of GINS-Pol epsilon on the replication origin.
Weaknesses:
The biochemical analysis should be carefully evaluated with more quantitative ways to strengthen the authors' conclusion.
We thank your positive assessment. We provided more quantitative information and tried to quantify the experiments as suggested (Please see the responses to [Recommendations for the authors]).
Recommendations for the authors:
Reviewer #1 (Recommendations for the authors):
I have several concerns that I will outline below, accompanied by my suggestions.
(1) "The title of the paper- "Structural and functional insights into Cdc45 recruitment by Sld7-Sld3 for CMG complex Formation," appears misleading because it appears that authors present a structure of Sld3-Sld7 in complex with Cdc45, which is not the case here. If authors can provide additional structures proving the function of this complex, then this title justifies it. Otherwise, I recommend making a title that justifies the presented work in its current form.
Following the comment, we change the title to “Sld3CBD-Cdc45 structural insights into Cdc45 recruitment for CMG complex formation”.
(2) In lines 70-72, where the authors mention the known structures of different proteins, intermediates, and complexes, I recommend including PDB IDs of the described structures and reference citations. This will help the readers to analyze what is missing in the pathway and why this structure is essential.
Following the comment, we added PBDIDs and references (P3/L72-74).
(3) The representation of Figure 1A is unclear and looks clumsy. If the structure were rotated in another orientation, where α8 and α9 would be displayed on the forward side, it would be more helpful to understand the complex forming regions by looking at the structure. Also, I recommend highlighting the α8 and α9 in a contrasting color to be easily visible and attract readers' attention. Similarly, it would also be helpful if DHAA1 would be shown in a different color.
Following the comment, we modified the Figure1 to show α8 and α9 of Sld3CBD and DHAA1 of Cdc45 clearly in revised version.
(4) Can authors add a supplementary figure showing the probability of disorderness of the α8 helix region in the Sld3? Also, highlight what region became ordered in their structure.
Yes, we have showed the disordered α8 helix region and highlight ordered α8 in the Sld3 in Figure S4 A.
(5) Can you compare the Cdc45 long distorted helix (Supplementary Figure 3B) in the Sld3-Cdc45 complex with the Xenoupus and drosophila Cdc45 from their CMG structures? Also, can the authors explain why this helix is destabilized in their structure but is relatively stable in another Cdc45 structure (in CMG and HuCdc45)?
We have checked all Cdc45 from published cryo-EM CMG structures, including Xenopus CMG-donson (8Q6O) and Drosophila CMG (6RAW), and all of them ordered the long helix in the CMG complex, whereas this long helix was disordered in the crystal structure of Sld3CBD-Cdc45 and Entamoeba histolytica Cdc45. The crystal packing around the long helix showed that it looks to be stabilized by crystal packing only in huCdc45, therefore we suggested that this long helix is detestable for crystallization.
(6) I recommend adding the following parameters to Supplementary Table 2: 1. Rmerge values, 2. Wilson B factor, 3. Average B factor, and 4. Total number of molecules in ASU.
We are sorry to make a mistake about Rmerge in Table 2. We correct it. We added the Wilson B factor, the average B factor, and the total number of Sld3CBD-Cde45 in ASU.
(7) Can authors provide the B factor values of the α8 helix of Sld3?
We checked the B factor values of the helix α8CTP of Sld3 in Sld3CBD-Cdc45. Since this helix binds to Cdc45 stably, the average B factor of the main chain is 45 Å2 less than that of the whole structure. We added the average B factor of helix α8CTP into the Supplementary Figure 4A legend.
(8) Can authors explain why higher Ramachandran outliers exist in their structure? Can it be reduced below 1% during refinement?
There are 13 outliers (1.67%) in different places: four are close to the disorder regions (poor electron map), four are in a loop with poor map and the remains are turn parts or a loop. For the residues with poor electron maps, we could not modify them to the allow Ramachandran region with low Rfree value, so we could not reduce them to below 1% during refinement while keeping the current Rfree value.
(9) In Supplementary Figure 8, please show the CD spectra of the Sld3WT. Why is the Sld3-3S peak relatively flat? Was the sample precipitating while doing the measurements, or does it have less concentration than others?
To check the folding of the mutants, we did CD experiments with the estimated secondary structure elements. Because WT Sld3CBD was prepared in a complex with Cdc45, while the mutants of Sld3CBD existed along, we calculated the elements of secondary structure from the crystal structure of Sld3CBD-Cdc45. The concentration of samples was controlled to the same level for CD measurement. The relative plat of the Sld3-3S peak may be caused by precipitating while doing the measurement.
(10) Can authors generate the alpha fold three models of the Sld3CBD-Cdc45-MCM-dsDNA and SCMG-dsDNA and compare them with the models they have generated?
We tried to predict the Sld3CBD-Cdc45-MCM-dsDNA and SCMG-dsDNA using Alphafold3. Although the results showed similar structures to our models, many parts were disordered. So, we did not use the predicted structures.
(11) The authors say that the overall molecular mass of the Sld7-Sld3ΔC-Cdc45 was >400kDa on the SEC column. However, the column used for purifying this complex and the standards that were run on it for molecular weight calculations have not been written anywhere. If the Superdex 200 column was used, then the sample of more than 400kDa should not elute at the position shown in Supplementary Figure 2B. I recommend showing the standard MW plot and where the elution volume of the Sld7-Sld3ΔC-Cdc45 lies on the standard curve. Also, add how molecular weight calculations were done and the calculated molecular mass.
Following the comment, we added a measurement of Superdex 200 16/60 column (SEC) using a standard sample kit into Supplementary Figure 2 to show that the molecular weight of the peak at the position was estimated to be > 400 k Da.
(12) I also recommend using at least one of the techniques, either SEC-MALS or AUC, to calculate the actual molecular mass of the Sld7-Sld3ΔC-Cdc45 complex and to find its oligomeric state. If the authors want to prove their hypothesis that a dimer of this complex binds to MCMDH, it is essential to show that it exists as a dimer. Based on the current SEC profile, it appears as a monomer peak if the S200 SEC column is being used.
As the response to (11), we added the standard MW plot (measurement using Superdex 200 16/60 column) using a standard sample kit. The molecular weight at the peak elution position of Sld7-Sld3ΔC-Cdc45 was estimated to be 429k Da. Considering that the Sld7-Sld3ΔC-Cdc45 dimer should be a flexible long-shaped molecule, the elution position could be at a larger molecular weight position than the real one (158 x 2 k Da). We also tried to confirm the particle size using SEC-SAXS, as the response to the next question (13).
(13) Dynamic light scattering is not the most accurate method for calculating intermolecular distance. I recommend using another technique that calculates the accurate molecular distances between two Cdc45 if Sld7-Sld3ΔC-Cdc45 is forming a dimer. Techniques such as FRET could be used. Otherwise, some complementary methods, such as SAXS, could also be used to generate a low-resolution envelope and fit the speculated dimer model inside, or authors could try negative staining the purified Sld7-Sld3ΔC-Cdc45 and generate 2D class averages and low-resolution ab initio models to see how the structure of this complex appears and whether it satisfies the speculated model of the dimeric complex.
We have tried both negative staining TEM and SEC-SAXS experiments. We could not obtain images good enough of negative staining of TEM to generate 2D class averages and low-resolution ab initio models. The results of SEC-SAXS provided a molecular weight of 370 - 420 kDa, and an Rg > 85 Å, which are consistent with our conclusion from SEC and DLS results but with large error due to the measurement temperature at 10-15°C (measuring equipment limitation). The peak of SCE-SAXS under measurement conditions was not as sharp as purification at 4°C and SAXS data is not good enough to make a molecular model, so we did not add them to our manuscript.
(14) Authors mentioned in the introduction section (lines 72-73) that based on the single-molecule experiments, Cdc45 is recruited in a stepwise manner to MCMDH. If this is true and if Sld7-Sld3ΔC-Cdc45 forms a dimer, this is also true, then for stepwise recruitment, the dimer will have to break into monomers, and this will be an energy-expensive process for the cell. So, would such a process occur physiologically? Can the authors explain how this would physiologically happen inside the cell?
Sld7-Sld3-Cdc45 consists of domains linked by long loops, so the dimer Cdc45-Sld3-[Sld7]2-Sld3-Cdc45 is flexible long-sharp. Such a flexible dimer does not mean that two Cdc45 molecules must bind to MCM DH simultaneously and may bind to MCM DH by stepwise manner. The dimer formation of Sld7-Sld3-Cdc45 is advantageous for recruiting efficiently and saving energy. Moreover, our proposal of Cdc45-Sld3-[Sld7]2-Sld3-Cdc45 on MCM DH could be a stage during CMG formation in the cell. Following the comment, we added such descriptions (P7/L194, and P10/L276-279).
(15) Can authors show experimentally that a dimer of Sld7-Sld3ΔC-Cdc45 is binding to MCMDH and not a monomer in a stepwise fashion?
In our study, we provided experiments of particle size to show the dimer of Sld7-Sld3-Cdc45 off MCM DH and a model of SCMG to indicate the dimer of Sld7-Sld3ΔC-Cdc45 on MCM DH. This question should be addressed future by the Cryo-EM of Sld7-Sld3-Cdc45-MCM DH or Sld7-Sld3-CMG. As the response to Q14, the flexible dimer of Sld7-Sld3ΔC-Cdc45 binding on MCMDH does not contradict the stepwise-loading fashion. The dimer of Sld7-Sld3ΔC-Cdc45 binding on MCM DH shows a stage.
(16) Can authors highlight where Sld7 will lie on their model shown in Figures 3A and 3C, considering their model shown in 3B is true?
We predict that the Sld7-Sld3-Cdc45 should be in a dimer form of Cdc45-Sld3-[Sld7]2-Sld3-Cdc45 based on the structures and the particle size analysis. The Sld7 dimer could be across MCM DH on the top of Figure 3A right and 3C right. However, we could not add the Sld7 molecule to the models because there is no interaction data between Sld7 and MCM.
(17) In Supplementary Figure 10, can authors show the residues between the loop region highlighted in the dotted circle to show that there is no steric clash between the residues in that region of their predicted model?
Following the comment, we added the residues in Supplementary Figure 10 (Supplementary Figure 11 in the revised version) to show no steric clash in our predicted model.
(18) It is essential to show experimentally that Sld3CBD neighbors MCM2 and binds Cdc45 on the opposite side of the GINS binding site. I recommend that the authors design an experiment that proves this statement. Mutagenesis experiments for the predicted residues that could be involved in interaction with proper controls might help to prove this point. Since this is the overall crux of the paper, it has to be demonstrated experimentally.
We thank the reviewer’s recommendation. Our structural analysis experiment shows the interaction information between Sld3CBD and Cdc45 at 2.6 Å resolution. The Sld3CBD-binding site, GINS-binding site, and MCM-binding site of Cdc45 are completely different, indicating that the Sld3CBD, Cdc45 and GINS could bind to MCM together. The SCMG model confirmed such a binding manner. Following the recommendation, we added mutant analysis of Cdc45 G367D and W481R, which was reported to disrupt the binding to MCM and GINS, respectively. Both mutants do not affect the binging to Sld3CBD as we predicted (Supplementary Figure 9B). We modified our manuscript and discussed this point more clearly (P7/L170-173).
(19) I recommend rewriting the sentence in lines 208-210. During EMSA experiments, new bands do not appear; instead, there is no shift at lower ratios, so you see a band similar to the control for Sld3CBD-Cdc45. So, re-write the sentence correctly to avoid confusion when interpreting the result.
Following the comment, we rewrote this sentence to "The ssDNA band remained (Figure 4B) and new bands corresponding to the ssDNA–protein complex appeared in CBB staining PAGE (Supplementary Figures 13) when the Sld3CBD–Cdc45 complex was mixed with ssDNA at the same ratio, indicating that the binding affinity of Sld3CBD–Cdc45 for ssDNA was lower than that of Sld3CBD alone” (P8/L226-229)
(20) Since CDK-mediated phosphorylation of Sld3 is known to be required for GINS loading, the ssDNA binding affinity of phosphorylated Sld3 remains the same. I wonder what would happen if phosphorylated Sld3 were used for the experiment shown in Figure 4B.
The CDK phosphorylation site is located at Sld3CTD and our ssDNA-binding experiment did not include the Sld3CTD, so phosphorylated Sld3 does not affect the results shown in Figure 4B.
(21) Sld3CBD-Cdc45 has a reduced binding affinity for ss DNA, and Sld7-Sld3ΔC-Cdc45 and Sl7-Sld3ΔC have a similar binding affinity to Sld3CBD based on figure 4B. It appears that Sld3CBD reduces the DNA binding affinity of CDC45 or vice versa. Is it correct to say so?
Our opinion is “vice versa”. Cdc45 reduces the ssDNA-binding affinity of Sld3CBD. Although we could not point out the ssDNA-binding sites of Sld3CBD, the surface charge of Sld3CBD implies that α8CTP could contribute to ssDNA-binding (Supplementary Figures 15).
(22) Cdc45 binds to the ssDNA by itself, but in the case of Sld3CBD-Cdc45, the binding affinity is reduced for Sld3CBD and Cdc45. Based on their structure, can authors explain what leads to this complex's reduced binding affinity to the ssDNA? Including a figure showing how Sld7-Sld3CBD-Cdc45 interacts with the DNA would be a nice idea.
Previous studies showed that Cdc45 binds tighter to long ssDNA (> 60 bases) and the C-terminus of Cdc45 is responsible for the ssDNA binding activity. The structure of Sld3CBD-Cdc45 shows the C-terminal domain DHHA1 of Cdc45 binds to Sld3CBD, which may lead to Sld3CBD-Cdc45 complex reduced ssDNA-binding affinity of Cdc45. We agree that showing a figure of how Sld7-Sld3CBD-Cdc45 interacts with ssDNA is a nice idea. However, there is no detailed interaction information between Sld7-Sld3Δ-Cdc45 and ssDNA, so we could not give a figure to show the ssDNA-binding manner. We added a figure to show the surface charges of Sld3CBD of Sld3CBD-Cdc45, and Sld3NTD-Sld7NTD, respectively (Supplemental Figure 15).
(23) Based on the predicted model of Sld7-Sld3 and Cdc45 complex, can authors explain how Sld7 would restore the DNA binding ability of the Sld3CBD?
It can be considered that Sld7 and Sld3NTD could bind ssDNA. Although we did not perform the ssDNA-binding assay of Sld7, the Sld3NTD-Sld7NTD surface shows a large positive charge area which may contribute to ssDNA-binding (Supplemental Figure 15). We added the explanation (P9/L245-248).
(24) It would be important to show binding measurements and Kd values of all the different complexes shown in Figure 4B with ssDNA to explain the dissociation of Cdc45 from Sld7-Sld3 after the CMG formation. I also recommend describing the statement from lines 224-227 more clearly how Sld7-Sld3-Cdc45 is loading Cdc45 on CMG.
As the reviewer mentioned, the binding measurements and Kd of values of all the different complexes are important to explain the dissociation of Sld7-Sld3 from CMG. The pull-down assay using chromatography may be affected by balancing the binding affinity and chromatography conditions. Therefore, we used EMSA with native-PAGE, which is closest to the natural state. However, the disadvantage is that the Kd values could not be estimated. For lines 224-227, the ssARS1-binding affinity of Sld3 and its complex should relate to the dissociation of Sld7–Sld3 from the CMG complex but not Cdc45 loading, because ssARS1 is unwound from dsDNA by the CMG complex after Cdc45 and GINS loading. We modified the description (P9/L248-251).
(25) Can authors explain why SDS-PAGE was used to assess the ssDNA (See line 420)?
We are sorry for making this mistake and corrected it to “polyacrylamide gel electrophoresis”.
(26) In line 421, can the authors elaborate on a TMK buffer?
We are sorry for this omission and added the content of the TMK buffer (P16/L453).
(27) I am curious to know if the authors also attempted to Crystallize the Sld7-Sld3CBD-Cdc45 complex. This complex structure would support the authors' hypothesis in this article.
We tried to crystallize Sld7-Sld3Δ-Cdc45 but could not get crystals. We also tried using cryo-EM but failed to obtain data.
Reviewer #2 (Recommendations for the authors):
(1) The manuscript would be strengthened if the authors acknowledged in greater detail how their work agrees with or disagrees with Itou et al. (PMID: 25126958 DOI: 10.1016/j.str.2014.07.001). The introduction insufficiently described the findings of that previous work in lines 63-64.
We compared Sld3CBD in Sld3CBD-Cdc45 to the monomer reported by Itou et al. (PMID: 25126958 DOI: 10.1016/j.str.2014.07.001) in the section of [The overall structure of Sld3CBD-Cdc45] and point out the structural similarity and difference (P5/L105-106), especially, conformation change of Sld3CBD α8 for binding to Cdcd45, which agrees to the mutant experiments of Itou et al., (P3/L126-127). Another Cdc45-binding site of Sld3CBD in the Sld3CBD-Cdc45 complex is α9 not residues predicted in previous studies.
(2) Figure 2. Could you please perform and present data from multiple biological replicates (e.g., at least two independent experiments) for each mutant strain? This would help ensure that the observed pull-downs (2A-B) and growth patterns (2C) are consistent and reproducible.
We have done pull-downs three times from co-expression to purification and pull-down assay. We added descriptions to the method of [Mutant analysis of Sld3 and Cdc45]. The growth patterns are two times in Figure 2C.
(3) Figure 3B. The match between the predicted complex length and particle size measured by dynamic light scattering (DLS) is striking. Did the authors run the analysis with vehicle controls and particle size standards? There is no mention of these controls.
Following the comment, we added the control data of buffer and standard protein lysozyme, and the descriptions to the method of [Dynamic light scattering].
(4) Figure 4. In lines 216-217, the authors write that the binding of the K. marxianus complex "demonstrates that the presence of Sld7 could restore the single-stranded DNA binding capacity of Sld3." Another explanation is that complexes from each species bind differently. If the authors want to make a strong claim, they should compare the binding of complexes containing the same proteins.
Agree with the comment, to make a strong claim using samples from the same source is better. Due to limitations in protein overexpression, we used Sld7-Sld3ΔC-Cdc45 from different sources two sources belong to the identical family (Saccharomycetaceae) and the proteins Sld7, Sld3 and Cdc45 have sequence conservation with similar structures (RMSD = 0.356, 1.392, and 0.891 for Ca atoms of Sld7CTD, Sld7NTD-Sld3NTD, and Sld3CBD-Cdc45) predicted by the alphafold3. Such similarity in source and protein level allows us to do the comparison. Moreover, we modified the description to “indicates that the presence of Sld7 and Sld3NTD could increase the ssDNA-binding affinity to a level comparable to that of Sld3CBD.
(5) The logic of the following is unclear: "Considering that ssDNA is unwound from dsDNA by the helicase CMG complex, Sld7-Sld3ΔC-Cdc45, and Sld7-Sld3C having a stronger ssDNA-binding capacity than Sld3CBD-Cdc45 may imply a relationship between the dissociation of Sld7-Sld3 from the CMG complex and binding to ssDNA unwound by CMG." (Lines 224-227). How do the authors imagine that the binding affinity difference due to Sld7 contributes to the release of Sld3? Please explain.
Considering that ssARS1 is unwound from dsARS1 by the activated helicase CMG complex formed after loading Cdc45 and GINS, Sld3–Sld7 having a stronger ssARS1-binding affinity may provide an advantage for the dissociation of Sld7–Sld3 from the CMG complex. We modified the sentence of Lines 224-227 (P9/L248-251).
(6) The authors suggest that the release of Sld3 from the helicase is related to its association with single-stranded ARS1 DNA. They refer to the work of Bruck et al. (doi: 10.1074/jbc.M111.226332), which demonstrates that single-stranded origin DNA inhibits the interaction between Sld3 and MCM2-7 in vitro. The authors selectively choose data from this previous work, only including data that supports their model while disregarding other data. This approach hinders progress in the field. Specifically, Bruck proposed a model in which the association of Sld3 and GINS with MCM2-7 is mutually exclusive, explaining how Sld3 is released upon CMG assembly. In Figure 3 of the authors' model, they suggest that Sld3 can associate with MCM2-7 through CDC45, even when GINS is bound. Furthermore, Bruck's work showed that ssARS1-2 does not disrupt the Sld3-Cdc45 interaction. Instead, Bruck's data demonstrated that ssARS1-2 disrupts the interaction between MCM2-7 and Sld3 without Cdc45. While we do not expect the authors to consider all data in the literature when formulating a model, we urge them to acknowledge and discuss other critical data that challenges their model. Additionally, it would be beneficial for the field if the authors include both modes of Sld3 interaction with MCM2-7 (i.e., directly with MCM or through CDC45) when proposing a model for how CMG assembly and Sld3 release occurs.
In our discussion, we referred to the studies of Bruck’s data (doi: 10.1074/jbc.M111.226332) but did not discuss more because we didn’t perform similar experiments in vitro, and we do not think that no discussion hinders progress in the field. Promoting research progress, the new experiment should provide a new proposal and updated knowledge. Although we do not know exactly the positional relationship between Sld3 and Dpb11-Sld2 on MCM during GINS recruiting, the Sld3CBD-Cdc45 structure shows clearly that the Sld3CBD-binding site of Cdc45 is completely different from that of GINS and MCM binding to Cdc45. The model SCMG confirmed such a binding manner, Sld3, Cdc45 and GINS could bind together. The competition of Sld3 and GINS for binding to Cdc45 or Cdc45-MCM reported by Bruck et. al, may be caused by the conformation change of Cdc45 DHHA1 between Sld3CBD-Cdc45 and CMG, or without other initiation factors (CMG formation is regulated by the initial factors). We modified the discussion (P10/L282-286). Regarding ssARS1-binding, we did not discuss with Bruck's data that ARS1-2 does not disrupt the Sld3-Cdc45 interaction, because the data does not conflict with our proposal, although the data does not have an advantage. We propose that the release of Sld3 and Sld7 from CMG could be associated with the binding of ssARS1 unwound by CMG, but the dissociation event of Sl3-Sld7 doesn’t only ssARS1-binding. The exploration of unwound-ssARS1 causes the conformation change of CMG, which may be another event for Sld3-Sld7 dissociation. However, we do not have more experiments to confirm this and Bruck’s ssDNA-binding experiment did not use all of Sld3, Cdc45 and MCM, so we do not discuss more with Bruck’ data in the revised version (P11/L303-305).,
Reviewer #3 (Recommendations for the authors):
Major points:
(1) Figure 1, Sld3CBD-Cdc45 complex: Please indicate the number of critical residues and those of alpha-helixes and beta-sheets in this Figure or Supplemental Figure to confirm the authors' claim.
Following the comment, we added the number of alpha-helixes and beta-sheets with residue numbers in Figure 1, and Supplemental Figures 4 and 5. We also added a topology diagram (Supplemental Figure 3).
(2) Figure 2A and B: Please quantify the interaction here with a proper statistical comparison.
In the experiments of Figures 2A and 2B, we used a co-expression system to co-purify the complexes and check their binding. For quantifying, we added the concentrations of the samples used in the Method of [Mutant analysis of Sld3 and Cdc45].
(3) Figure 3B, EMSA: If these are from the EMSA assay, at least free DNAs and protein-bound DNAs are present on the gel. However, the authors showed one band, which seems to be free DNA in Figure 3B and separately the smear band of the protein complex in Supplementary Figure 12, and judged the DNA binding by the disappearance of the band (line 207). Interestingly, in the case of Sld3CBD, there are few smear bands (Supplementary Figure 12). Where is DNA in this case? The disappearance could be due to the contaminated nucleases (need a control non-specific DNA). Without showing the Sld3CBD-DNA complex in the gel, the conclusion that the DNA binding activity of Sld3CBD-Cdc45 to DNA is lower than Sld3CBD alone (line 210) is very much speculative. The same is true for Sld7-Sld3dC-Cdc45.
Please explain the method (EMSA) briefly in the main text and show a whole gel in both Figures. If the authors insist that the Sld3 DNA-binding activity is altered with Cdc43 (and MCM), it is better to perform a more quantitative DNA binding assay such as BIAcore (surface plasmon), etc.
In the EMSA, we use SYBR (Figure 4B) and CBB (Supplementary Figure 13) staining to show bands of ssDNA and protein, respectively. As the reviewer mentioned, the disappearance of the bands could be due to the contaminated nucleases, we did experiments with non-specific ssDNA-binding as a control using the same proteins shown in Supplementary Figure 14. So, we are convinced that the disappearance of the ssDNA bands or not disappearance could occur when binding to protein or not. We added such explanations in the text (P9/L242-244). As we mentioned in the legend of Supplementary Figure 13, the Sld3CBD could not enter the gel, even when bound to ssDNA, because the pI values exceeded the pH of the running buffer.
Following the reviewer's comments, we attempted a pull-down experiment using Histag (C-terminal histag of Sld3CBD/Sld3ΔC). Unfortunately, we encountered difficulties in achieving the balance between binding and chromatography conditions.
(4) Figure 3B: Please quantify the DNA binding here with a proper statistical comparison with triplicate.
For EMSA (Figure 3B), we used samples of ssDNA:protein= 1:0. 1:1, 1:2, 1:4 and 0:1 molecular ratios with 10 pM as a 1 unit. We added concentrations of the samples in the Method of [Electrophoretic mobility shift assay for ssDNA binding].
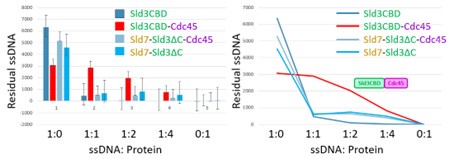
Following the comment, we tried to quantify the binding strength by integrating the grayscale of the bands in gel photos. However, we are concerned because this quantitative calculation through grayscale could not provide an accurate representation of results. Many sample groups cannot be run on one gel. Therefore, the gel differences in parameters cause large errors in the calculation as shown in Author response image 1. Although the calculated integral grayscale chart is consistent with our conclusion, we do not want to add this to our manuscript.
Author response image 1.
(5) Because of poor writing, the authors need to ask for English editing.
We are very sorry for the language. We asked a company (Editag, https:www.editage.jp) to do a native speaker revision and used AI to recheck English.
Minor points:
(1) Lines 47-58, Supplementary Figure 1: Although the sentences describe well how CMG assembles on the replication origin, the figure does not reflect what is written, but rather shows a simple schematic figure related to the work. However, for the general readers, it is very useful to see a general model of the CMG assembly. Then, the authors need to emphasize the steps focused in this study.
Thank you for your thoughtful comments. We optimized Figure 1 and hope it will be more understandable to general readers.
(2) Line 50, DDK[6F0L](superscript): what is 5F0L?
We are sorry for this mistake, that is a PDBID of the DDK structure. we deleted 6F0L.
(3) Lines 68 and 69, ssDNA and dsDNA: should be "single-stranded DNA (ssDNA)" and double-stranded DNA (dsDNA) when these words appear for the first time.
Following the comment, we modified it to “single-stranded DNA (ssDNA)” and “double-stranded DNA (dsDNA)” (P3/L68,70).
(4) Line 84, Cdc45s: What "s" means here?
We are sorry for this mistake, we modified it to “Cdc45”.
(5) Line 87, Sld3deltaC: What is Sld3deltaC? This is the deletion of either the Cdc45-binding domain or the C-terminal domain.
Sld3ΔC is a deletion of the C-terminal domain of Sld3. We added the residue range and explanation (P4/L91).
(6) Line 103: Although the authors mentioned beta-sheets 1-14 in the text, there is no indication in Figures. It is impossible to see the authors' conclusion.
The secondary structure elements of Sld3CBD-Cdc45 are shown in Supplementary Figures 4 and 5. Following the comment, we added a topology diagram of Sld3CBD and Cdc45 in the Sld3CBD-Cdc45 complex as Supplementary Figure 3 and added citations when describing structural elements.
(7) Line 106, huCdc45: Does this mean human Cdc45? If so, it should be "human CDC45 (huCDC45). CMG form is from budding yeast? Please specify the species.
Yes, huCdc45 is human Cdc45. We modified it into “human CDC45 (huCdc45)”.
(8) Line 107, Supplemental Figure 3B, black ovals: Please add "alpha7" in the Figure.
Following the comment, we added a label of Cdc45 α7 to Supplemental Figure 3B and 3C (Supplemental Figure 4B and 4C in revised version).
(9) Line 128, DHHA1: What is this? Please explain it in the text.
Following the comment, we added the information on DHHA1 (P3/L75-77).
(10) Line 130, beta13, and beta14: If the authors would like to point out these structures, please indicate where these sheets are in Figures.
We added a topology diagram as Supplementary Figure 3 to show the β-sheet in DHH and added a citation in the text.
(11) Line 133: Please add (Figure 1B) after the a8CTP.
Following the comment, we added “(Figure 1C)” (1B is 1C in revised version) after the α8CTP (P6/L133).
(12) Line 140: After DHHA1, please add (Figure 1C).
Following the comment, we added the figure citation after the DHHA1 (P6/L140).
(13) Line 142: After DHHA1, please add (Figure 1D).
Following the comment, we added the figure citation after the DHHA1 (P6/L142).
(14) Line 149, Sld3-Y seemed to retain a faint interaction with Cdc45. The Cdc45 band is too faint here. Moreover, as shown above, without the quantification with proper statistics, it is hard to draw this kind of conclusion.
We agree that the Cdc45 band corresponding to Sld3-Y in the pull-down assay was very faint, so we performed an in vivo experiment (Fig2C) to confirm this result.
(15) Line 149, Figure 2A and B: What kind of interaction assay was used here? Simple pull-down. It seems to eluate from the column. If so, how do the authors evaluate the presence of the proteins in different fractions? Please explain the method briefly in the main text.
Figure 2 shows a co-express pull-down binding assay. To describe the co-express pull-down experiments clearly, we added more explanations in the Methods [Mutation analysis of Sld3 and Cdc45].
(16) Line 154-155: Please show the quantification to see if the reduced binding is statistically significant.
Here, we explain why Cdc45-A remained Sld3CBD-bind ability. Although mutant Cdc45-A has reduced three hydrogen bonds with D344 of Sld3CBD, the remaining hydrogen-bond network keeps contact between Sld3CBD and Cdc45.
(17) Line 158, cell death: "No growth" does not mean cell death. Please rephrase here.
Following the comment, we modified it to “no growth” (P6/L158).
(18) Line 166: After CMG dimer, please add "respectively".
Following the comment, we added the word “, respectively” after CMG dimer (P7/L178).
(19) Line 194-195: I can not catch the meaning. Please rephrase here to clarify the claim. What are ssARS1-2 and ARS1-5?
Following the comment, we added more information about ssDNA fragments at the beginning of this section (P8/L210-214).
(20) Figure 4A and Supplemental Figure 12 top, schematic figure of ARS region. It is hard to catch. More explanation of the nature of the DNA substrates and much better schematic presentations would be appreciated.
Following the comment, we added more information about ARS1 to the figure legend.
(21) Figure 1A, dotted ovals should be dotted squares as shown in the enlarged images on the bottom.
Following the comment, we modified Figure 1A and the legend to change the dotted ovals into dotted squares.
-

-

-

eLife Assessment
This valuable paper describes the crystal structure of a complex of Sld3-Cdc45-binding domain (CBD) with Cdc45, which is essential for the assembly of an active Cdc45- MCM-GINS (CMG) double hexamers at the replication origin. Although the results shown in the paper are of interest to researchers in DNA replication and genome stability, the biochemical analysis of protein-protein interaction and DNA binding is incomplete, and the paper needs additional data and revised discussion.
-

Reviewer #1 (Public review):
Summary:
The crystal structure of the Sld3CBD-Cdc45 complex presented by Li et al. is a novel contribution that significantly advances our understanding of CMG formation during the rate-limiting step of DNA replication initiation. This structure provides insights into the intermediate steps of CMG formation. The study builds upon previously known structures of Sld3 and Cdc45 and offers new perspectives into how Cdc45 is loaded onto MCM DH through Sld3-Sld7. The most notable finding is the structural difference in Sld3CBD when bound to Cdc45, particularly the arrangement of the α8-helix, which is essential for Cdc45 binding and may also pertain to its metazoan counterpart, Treslin. Additionally, the conformational shift in the DHHA1 domain of Cdc45 suggests a possible mechanism for its binding to MCM2NTD.
Stre…
Reviewer #1 (Public review):
Summary:
The crystal structure of the Sld3CBD-Cdc45 complex presented by Li et al. is a novel contribution that significantly advances our understanding of CMG formation during the rate-limiting step of DNA replication initiation. This structure provides insights into the intermediate steps of CMG formation. The study builds upon previously known structures of Sld3 and Cdc45 and offers new perspectives into how Cdc45 is loaded onto MCM DH through Sld3-Sld7. The most notable finding is the structural difference in Sld3CBD when bound to Cdc45, particularly the arrangement of the α8-helix, which is essential for Cdc45 binding and may also pertain to its metazoan counterpart, Treslin. Additionally, the conformational shift in the DHHA1 domain of Cdc45 suggests a possible mechanism for its binding to MCM2NTD.
Strengths:
The manuscript is generally well-written, with a precise structural analysis and a solid methodological section that will significantly advance future studies in the field. The predictions based on structural alignments are intriguing and provide a new direction for exploring CMG formation, potentially shaping the future of DNA replication research.
Weaknesses:
The main weakness of the manuscript lies in the lack of experimental validation for the proposed Sld3-Sld7-Cdc45 model. Specifically, the claim that Sld3 binding to Cdc45-MCM does not inhibit GINS binding, a finding that contradicts previous research, is not sufficiently substantiated with experimental evidence. To strengthen their model, the authors must provide additional experimental data to support this mechanism. Also, the authors have not compared the recently published Cryo-EM structures of the metazoan CMG helicases with their predicted models to see if Sld3/Treslin does not cause any clash with the GINS when bound to the CMG. Still, the work holds great potential in its current form but requires further experiments to confirm the authors' conclusions.
-

Reviewer #2 (Public review):
Summary
The manuscript presents valuable findings, particularly in the crystal structure of the Sld3CBD-Cdc45 interaction and the identification of additional sequences involved in their binding. The modeling of the Sld7-Sld3CBD-CDC45 subcomplex is novel, and the results provide insights into potential conformational changes that occur upon interaction. However, the work remains incomplete as several main claims are only partially supported by experimental data, particularly the proposed model for Sld3 interaction with GINS on the CMG. Additionally, the single-stranded DNA binding data from different species do not convincingly advance the manuscript's central arguments.
Strengths
(1) The Sld3CBD-Cdc45 structure is a novel contribution, revealing critical residues involved in the interaction.
(2) The model …
Reviewer #2 (Public review):
Summary
The manuscript presents valuable findings, particularly in the crystal structure of the Sld3CBD-Cdc45 interaction and the identification of additional sequences involved in their binding. The modeling of the Sld7-Sld3CBD-CDC45 subcomplex is novel, and the results provide insights into potential conformational changes that occur upon interaction. However, the work remains incomplete as several main claims are only partially supported by experimental data, particularly the proposed model for Sld3 interaction with GINS on the CMG. Additionally, the single-stranded DNA binding data from different species do not convincingly advance the manuscript's central arguments.
Strengths
(1) The Sld3CBD-Cdc45 structure is a novel contribution, revealing critical residues involved in the interaction.
(2) The model structures generated from the crystal data are well presented and provide valuable insights into the interaction sequences between Sld3 and Cdc45.
(3) The experiments testing the requirements for interaction sequences are thorough and conducted well, with clear figures supporting the conclusions.
(4) The conformational changes observed in Sld3 and Cdc45 upon binding are interesting and enhance our understanding of the interaction.
(5) The modeling of the Sld7-Sld3CBD-CDC45 subcomplex is a new and valuable addition to the field.
Weaknesses
(1) The proposed model for Sld3 interacting with GINS on the CMG needs more experimental validation and conflicts with published findings. These discrepancies need more detailed discussion and exploration.
(2) The section on the binding of Sld3 complexes to origin single-stranded DNA needs significant improvement. The comparisons between Sld3-CBD, Sld3CBD-Cdc45, and Sld7-Sld3CBD-Cdc45 involve complexes from different species, limiting the comparisons' value.
(3) The authors' model proposing the release of Sld3 from CMG based on its binding to single-stranded DNA is unclear and needs more elaboration.
-

Reviewer #3 (Public review):
Summary:
The paper by Li et al. describes the crystal structure of a complex of Sld3-Cdc45-binding domain (CBD) with Cdc45 and a model of the dimer of an Sld3-binding protein, Sld7, with two Sld3-CBD-Cdc45 for the tethering. In addition, the authors showed the genetic analysis of the amino acid substitution of residues of Sld3 in the interface with Cdc45 and biochemical analysis of the protein interaction between Sld3 and Cdc45 as well as DNA binding activity of Sld3 to the single-strand DNAs of the ARS sequence.
Strengths:
The authors provided a nice model of an intermediate step in the assembly of an active Cdc45-MCM-GINS (CMG) double hexamers at the replication origin, which is mediated by the Sld3-Sld7 complex. The dimer of the Sld3-Sld7 complexes tethers two MCM hexamers together for the recruitment of …
Reviewer #3 (Public review):
Summary:
The paper by Li et al. describes the crystal structure of a complex of Sld3-Cdc45-binding domain (CBD) with Cdc45 and a model of the dimer of an Sld3-binding protein, Sld7, with two Sld3-CBD-Cdc45 for the tethering. In addition, the authors showed the genetic analysis of the amino acid substitution of residues of Sld3 in the interface with Cdc45 and biochemical analysis of the protein interaction between Sld3 and Cdc45 as well as DNA binding activity of Sld3 to the single-strand DNAs of the ARS sequence.
Strengths:
The authors provided a nice model of an intermediate step in the assembly of an active Cdc45-MCM-GINS (CMG) double hexamers at the replication origin, which is mediated by the Sld3-Sld7 complex. The dimer of the Sld3-Sld7 complexes tethers two MCM hexamers together for the recruitment of GINS-Pol epsilon on the replication origin.
Weaknesses:
The biochemical analysis should be carefully evaluated with more quantitative ways to strengthen the authors' conclusion.
-

Author response:
Reviewer #1 (Public review):
Summary:
The crystal structure of the Sld3CBD-Cdc45 complex presented by Li et al. is a novel contribution that significantly advances our understanding of CMG formation during the rate-limiting step of DNA replication initiation. This structure provides insights into the intermediate steps of CMG formation. The study builds upon previously known structures of Sld3 and Cdc45 and offers new perspectives into how Cdc45 is loaded onto MCM DH through Sld3-Sld7. The most notable finding is the structural difference in Sld3CBD when bound to Cdc45, particularly the arrangement of the α8-helix, which is essential for Cdc45 binding and may also pertain to its metazoan counterpart, Treslin. Additionally, the conformational shift in the DHHA1 domain of Cdc45 suggests a possible mechanism for its …
Author response:
Reviewer #1 (Public review):
Summary:
The crystal structure of the Sld3CBD-Cdc45 complex presented by Li et al. is a novel contribution that significantly advances our understanding of CMG formation during the rate-limiting step of DNA replication initiation. This structure provides insights into the intermediate steps of CMG formation. The study builds upon previously known structures of Sld3 and Cdc45 and offers new perspectives into how Cdc45 is loaded onto MCM DH through Sld3-Sld7. The most notable finding is the structural difference in Sld3CBD when bound to Cdc45, particularly the arrangement of the α8-helix, which is essential for Cdc45 binding and may also pertain to its metazoan counterpart, Treslin. Additionally, the conformational shift in the DHHA1 domain of Cdc45 suggests a possible mechanism for its binding to MCM2NTD.
Strengths:
The manuscript is generally well-written, with a precise structural analysis and a solid methodological section that will significantly advance future studies in the field. The predictions based on structural alignments are intriguing and provide a new direction for exploring CMG formation, potentially shaping the future of DNA replication research.
Weaknesses:
The main weakness of the manuscript lies in the lack of experimental validation for the proposed Sld3-Sld7-Cdc45 model. Specifically, the claim that Sld3 binding to Cdc45-MCM does not inhibit GINS binding, a finding that contradicts previous research, is not sufficiently substantiated with experimental evidence. To strengthen their model, the authors must provide additional experimental data to support this mechanism. Also, the authors have not compared the recently published Cryo-EM structures of the metazoan CMG helicases with their predicted models to see if Sld3/Treslin does not cause any clash with the GINS when bound to the CMG. Still, the work holds great potential in its current form but requires further experiments to confirm the authors' conclusions.
We appreciate the reviewers’ careful reading and the comments.
The structure of Sld3CBD-Cdc45 showed that the binding site of Cdc45 to Sld3CBD was distinct from the binding ranges of Cdc45 to GINS and MCM, indicating that the Sld3CBD, MCM, and GINS bind to separate sites of Cdc45 on the CMG complex. The SCMG-DNA model confirmed such a binding situation but did not show whether the binding of Sld3 to Cdc45 affects the recruitment of GINS (by GINS-Dbp11-Sld2) for CMG formation. We will modify our manuscript and discuss this point. Also, we will check the recently published Cryo-EM structures of the metazoan CMG helicases with their predicted models to confirm our conclusions. We will try to conduct the experiments as suggested.
Reviewer #2 (Public review):
Summary
The manuscript presents valuable findings, particularly in the crystal structure of the Sld3CBD-Cdc45 interaction and the identification of additional sequences involved in their binding. The modeling of the Sld7-Sld3CBD-CDC45 subcomplex is novel, and the results provide insights into potential conformational changes that occur upon interaction. However, the work remains incomplete as several main claims are only partially supported by experimental data, particularly the proposed model for Sld3 interaction with GINS on the CMG. Additionally, the single-stranded DNA binding data from different species do not convincingly advance the manuscript's central arguments.
Strengths
(1) The Sld3CBD-Cdc45 structure is a novel contribution, revealing critical residues involved in the interaction.
(2) The model structures generated from the crystal data are well presented and provide valuable insights into the interaction sequences between Sld3 and Cdc45.
(3) The experiments testing the requirements for interaction sequences are thorough and conducted well, with clear figures supporting the conclusions.
(4) The conformational changes observed in Sld3 and Cdc45 upon binding are interesting and enhance our understanding of the interaction.
(5) The modeling of the Sld7-Sld3CBD-CDC45 subcomplex is a new and valuable addition to the field.
Weaknesses
(1) The proposed model for Sld3 interacting with GINS on the CMG needs more experimental validation and conflicts with published findings. These discrepancies need more detailed discussion and exploration.
(2) The section on the binding of Sld3 complexes to origin single-stranded DNA needs significant improvement. The comparisons between Sld3-CBD, Sld3CBD-Cdc45, and Sld7-Sld3CBD-Cdc45 involve complexes from different species, limiting the comparisons' value.
(3) The authors' model proposing the release of Sld3 from CMG based on its binding to single-stranded DNA is unclear and needs more elaboration.
We appreciate your positive comments. As suggested, we will try to improve the experiments and manuscript and discuss in more detail, including the interaction between Sld3 and GINS on the CMG, ssDNA-binding section, and the explanations of why we use different species for comparison and more elaboration on the Sld3-release proposal.
Reviewer #3 (Public review):
Summary:
The paper by Li et al. describes the crystal structure of a complex of Sld3-Cdc45-binding domain (CBD) with Cdc45 and a model of the dimer of an Sld3-binding protein, Sld7, with two Sld3-CBD-Cdc45 for the tethering. In addition, the authors showed the genetic analysis of the amino acid substitution of residues of Sld3 in the interface with Cdc45 and biochemical analysis of the protein interaction between Sld3 and Cdc45 as well as DNA binding activity of Sld3 to the single-strand DNAs of the ARS sequence.
Strengths:
The authors provided a nice model of an intermediate step in the assembly of an active Cdc45-MCM-GINS (CMG) double hexamers at the replication origin, which is mediated by the Sld3-Sld7 complex. The dimer of the Sld3-Sld7 complexes tethers two MCM hexamers together for the recruitment of GINS-Pol epsilon on the replication origin.
Weaknesses:
The biochemical analysis should be carefully evaluated with more quantitative ways to strengthen the authors' conclusion.
We thank your positive assessment. We will provide more quantitative information and try to quantify the experiments as suggested.
-