Deficiency of IQCH causes male infertility in humans and mice
Curation statements for this article:-
Curated by eLife
eLife assessment
This valuable study describes mice with a knock out of the IQ motif-containing H (IQCH) gene, to model a human loss-of-function mutation in IQCH associated with male sterility. While the evidence for interaction between IQCH and potential RNA binding proteins is limited, the human infertility is reproduced in the mouse, making it a compelling model. The paper could be of interest to cell biologists and male reproductive biologists working on the sperm flagellar cytoskeleton and mitochondrial structure.
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
IQ motif-containing proteins can be recognized by calmodulin (CaM) and are essential for many biological processes. However, the role of IQ motif-containing proteins in spermatogenesis is largely unknown. In this study, we identified a loss-of-function mutation in the novel gene IQ motif-containing H ( IQCH ) in a Chinese family with male infertility characterized by a cracked flagellar axoneme and abnormal mitochondrial structure. To verify the function of IQCH, Iqch knockout (KO) mice were generated via CRISPR-Cas9 technology. As expected, the Iqch KO male mice exhibited impaired fertility, which was related to deficient acrosome activity and abnormal structures of the axoneme and mitochondria, mirroring the patient phenotypes. Mechanistically, IQCH can bind to CaM and subsequently regulate the expression of RNA-binding proteins (especially HNRPAB), which are indispensable for spermatogenesis. Overall, this study revealed the function of IQCH, expanded the role of IQ motif-containing proteins in reproductive processes, and provided important guidance for genetic counseling and genetic diagnosis of male infertility.
Article activity feed
-

-

-

-

eLife assessment
This valuable study describes mice with a knock out of the IQ motif-containing H (IQCH) gene, to model a human loss-of-function mutation in IQCH associated with male sterility. While the evidence for interaction between IQCH and potential RNA binding proteins is limited, the human infertility is reproduced in the mouse, making it a compelling model. The paper could be of interest to cell biologists and male reproductive biologists working on the sperm flagellar cytoskeleton and mitochondrial structure.
-

Reviewer #3 (Public Review):
In this study, Ruan et al. investigate the role of the IQCH gene in spermatogenesis, focusing on its interaction with calmodulin and its regulation of RNA-binding proteins. The authors examined sperm from a male infertility patient with an inherited IQCH mutation as well as Iqch CRISPR knockout mice. The authors found that both human and mouse sperm exhibited structural and morphogenetic defects in multiple structures, leading to reduced fertility in Ichq-knockout male mice. Molecular analyses such as mass spectrometry and immunoprecipitation indicated that RNA-binding proteins are likely targets of IQCH, with the authors focusing on the RNA-binding protein HNRPAB as a critical regulator of testicular mRNAs. The authors used in vitro cell culture models to demonstrate an interaction between IQCH and …
Reviewer #3 (Public Review):
In this study, Ruan et al. investigate the role of the IQCH gene in spermatogenesis, focusing on its interaction with calmodulin and its regulation of RNA-binding proteins. The authors examined sperm from a male infertility patient with an inherited IQCH mutation as well as Iqch CRISPR knockout mice. The authors found that both human and mouse sperm exhibited structural and morphogenetic defects in multiple structures, leading to reduced fertility in Ichq-knockout male mice. Molecular analyses such as mass spectrometry and immunoprecipitation indicated that RNA-binding proteins are likely targets of IQCH, with the authors focusing on the RNA-binding protein HNRPAB as a critical regulator of testicular mRNAs. The authors used in vitro cell culture models to demonstrate an interaction between IQCH and calmodulin, in addition to showing that this interaction via the IQ motif of IQCH is required for IQCH's function in promoting HNRPAB expression. In sum, the authors concluded that IQCH promotes male fertility by binding to calmodulin and controlling HNRPAB expression to regulate the expression of essential mRNAs for spermatogenesis. These findings provide new insight into molecular mechanisms underlying spermatogenesis and how important factors for sperm morphogenesis and function are regulated.
The strengths of the study include the use of mouse and human samples, which demonstrate a likely relevance of the mouse model to humans; the use of multiple biochemical techniques to address the molecular mechanisms involved; the development of a new CRISPR mouse model; ample controls; and clearly displayed results. Assays are done rigorously and in a quantitative manner. Overall, the claims made by the authors in this manuscript are well-supported by the data provided.
-

Author response:
The following is the authors’ response to the previous reviews
It is unclear to us why you did not adjust the title to better reflect the well-supported claims of the paper, i.e., that this is a valuable model for human loss-of-function mutations in IQCH.
Thanks for the editor’s suggestion. We have changed the title to “Deficiency of IQCH causes male infertility in humans and mice.” Additionally, we have provided the original images of the gels or blots as a zipped folder.
-

-

Author response:
The following is the authors’ response to the previous reviews.
Public Reviews:
Reviewer #1 (Public Review):
By identifying a loss of function mutant of IQCH in infertile patient, Ruan et al. shows that IQCH is essential for spermiogenesis by generating a knockout mouse model of IQCH. Similar to infertile patient with mutant of IQCH, Iqch knockout mice are characterized by a cracked flagellar axoneme and abnormal mitochondrial structure. Mechanistically, IQCH regulates the expression of RNA-binding proteins (especially HNRPAB), which are indispensable for spermatogenesis.
Although this manuscript contains a potentially interesting piece of work that delineates a mechanism of IQCH that associates with spermatogenesis, this reviewer feels that a number of issues require clarification and re-evaluation for a better …
Author response:
The following is the authors’ response to the previous reviews.
Public Reviews:
Reviewer #1 (Public Review):
By identifying a loss of function mutant of IQCH in infertile patient, Ruan et al. shows that IQCH is essential for spermiogenesis by generating a knockout mouse model of IQCH. Similar to infertile patient with mutant of IQCH, Iqch knockout mice are characterized by a cracked flagellar axoneme and abnormal mitochondrial structure. Mechanistically, IQCH regulates the expression of RNA-binding proteins (especially HNRPAB), which are indispensable for spermatogenesis.
Although this manuscript contains a potentially interesting piece of work that delineates a mechanism of IQCH that associates with spermatogenesis, this reviewer feels that a number of issues require clarification and re-evaluation for a better understanding of the role of IQCH in spermatogenesis.
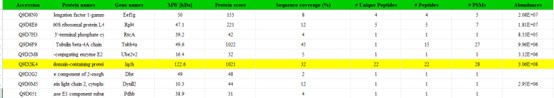
Line 251 - 253, "To elucidate the molecular mechanism by which IQCH regulates male fertility, we performed liquid chromatography tandem mass spectrometry (LC‒MS/MS) analysis using mouse sperm lysates and detected 288 interactors of IQCH (Figure 5-source data 1)."
The reviewer had already raised significant concerns regarding the text above, noting that "LC‒MS/MS analysis using mouse sperm lysates" would not identify interactors of IQCH. However, this issue was not addressed in the revised manuscript. In the Methods section detailing LC-MS/MS, the authors stated that it was conducted on "eluates obtained from IP". However, there was no explanation provided on how IP for LC-MS/MS was performed. Additionally, it was unclear whether LC-MS or LC-MS/MS was utilized. The primary concern is that if LC‒MS/MS was conducted for the IP of IQCH, IQCH itself should have been detected in the results; however, as indicated by Figure 5-source data 1, IQCH was not listed.
Thanks to reviewer’s comments. Additional details regarding the IP protocol for LC-MS/MS analysis have been included in the methods section in the revised manuscript. Furthermore, we apologize for the previous inconsistencies in the terminology used for LC-MS/MS and have now ensured its consistent usage throughout the document. Regarding the primary concern about the absence of IQCH in Figure 5-source data 1, our study only showed identifying proteins that interact with IQCH, not IQCH itself. Additionally, we conducted co-IP experiments to validate the interactions identified by LC-MS/MS analysis. Actually, we identified the IQCH itself by LC-MS/MS analysis (Author response table 1).
Author response table 1.
Results of the LC-MS/MS analysis.
Recommendations for the authors:
Reviewer #1 (Recommendations For The Authors):
The authors should know what experiments have been done for the studies.
We apologize for our oversights. The method for RNA-binding protein immunoprecipitation (RIP) has been detailed in the revised manuscript.
Typos still remain in the text, e.g., line 253, "Fiugre".
We are sorry for the spelling errors. We have engaged professional editing services to refine our manuscript.
-

eLife assessment
This valuable study describes mice with a knock out of the IQ motif-containing H (IQCH) gene, to model a human loss-of-function mutation in IQCH associated with male sterility. The infertility is reproduced in the mouse, making it a compelling model, but the mechanistic experiments provide only incomplete evidence for interaction between IQCH and potential RNA binding proteins, which are prominently mentioned in the title. The paper, which has undergone multiple rounds of review, could be of interest to cell biologists and male reproductive biologists working on the sperm flagellar cytoskeleton and mitochondrial structure.
-

Reviewer #3 (Public Review):
In this study, Ruan et al. investigate the role of the IQCH gene in spermatogenesis, focusing on its interaction with calmodulin and its regulation of RNA-binding proteins. The authors examined sperm from a male infertility patient with an inherited IQCH mutation as well as Iqch CRISPR knockout mice. The authors found that both human and mouse sperm exhibited structural and morphogenetic defects in multiple structures, leading to reduced fertility in Ichq-knockout male mice. Molecular analyses such as mass spectrometry and immunoprecipitation indicated that RNA-binding proteins are likely targets of IQCH, with the authors focusing on the RNA-binding protein HNRPAB as a critical regulator of testicular mRNAs. The authors used in vitro cell culture models to demonstrate an interaction between IQCH and …
Reviewer #3 (Public Review):
In this study, Ruan et al. investigate the role of the IQCH gene in spermatogenesis, focusing on its interaction with calmodulin and its regulation of RNA-binding proteins. The authors examined sperm from a male infertility patient with an inherited IQCH mutation as well as Iqch CRISPR knockout mice. The authors found that both human and mouse sperm exhibited structural and morphogenetic defects in multiple structures, leading to reduced fertility in Ichq-knockout male mice. Molecular analyses such as mass spectrometry and immunoprecipitation indicated that RNA-binding proteins are likely targets of IQCH, with the authors focusing on the RNA-binding protein HNRPAB as a critical regulator of testicular mRNAs. The authors used in vitro cell culture models to demonstrate an interaction between IQCH and calmodulin, in addition to showing that this interaction via the IQ motif of IQCH is required for IQCH's function in promoting HNRPAB expression. In sum, the authors concluded that IQCH promotes male fertility by binding to calmodulin and controlling HNRPAB expression to regulate the expression of essential mRNAs for spermatogenesis. These findings provide new insight into molecular mechanisms underlying spermatogenesis and how important factors for sperm morphogenesis and function are regulated.
The strengths of the study include the use of mouse and human samples, which demonstrate a likely relevance of the mouse model to humans; the use of multiple biochemical techniques to address the molecular mechanisms involved; the development of a new CRISPR mouse model; ample controls; and clearly displayed results. Assays are done rigorously and in a quantitative manner. Overall, the claims made by the authors in this manuscript are well-supported by the data provided.
-

-

Author response:
The following is the authors’ response to the previous reviews.
Public Reviews:
Reviewer #1 (Public Review):
Although this manuscript contains a potentially interesting piece of work that delineates a mechanism of IQCH that associates with spermatogenesis, this reviewer feels that a number of issues require clarification and re-evaluation for a better understanding of the role of IQCH in spermatogenesis. With the shortage of logics and supporting data, causal relationships are still not clear among IQCH, CaM, and HNRPAB. The most serious point in this manuscript could be that the authors try to generalize their interpretations with too simplified model from limited pieces of their data. The way the data and the logic are presented needs to be largely revised, and several interpretations should be supported by …
Author response:
The following is the authors’ response to the previous reviews.
Public Reviews:
Reviewer #1 (Public Review):
Although this manuscript contains a potentially interesting piece of work that delineates a mechanism of IQCH that associates with spermatogenesis, this reviewer feels that a number of issues require clarification and re-evaluation for a better understanding of the role of IQCH in spermatogenesis. With the shortage of logics and supporting data, causal relationships are still not clear among IQCH, CaM, and HNRPAB. The most serious point in this manuscript could be that the authors try to generalize their interpretations with too simplified model from limited pieces of their data. The way the data and the logic are presented needs to be largely revised, and several interpretations should be supported by direct evidence.
Response: Thank you for the reviewer’s comment. IQCH is a calmodulin-binding protein, and the binding of IQCH and CaM was confirmed by LC-MS/MS analysis and co-IP assay using sperm lysate. We thus speculated that if the interaction of IQCH and CaM might be a prerequisite for IQCH function. To prove that speculation, we took HNRPAB as an example. We knocked down IQCH in cultured cells, and a decrease in the expression of HNRPAB was observed. Similarly, when we knocked down CaM in cultured cells, and a decrease in the expression of HNRPAB was also detected. However, these results cannot exclude that IQCH or CaM could regulate HNRPAB expression alone. To investigate that if IQCH or CaM could regulate HNRPAB expression alone, we overexpressed IQCH in cells that knocked down CaM, while the expression of HNRPAB cannot be rescued, suggesting that IQCH cannot regulate HNRPAB expression when CaM is reduced. In consistent, we overexpressed CaM in cells that knocked down IQCH, while the expression of HNRPAB cannot be rescued, suggesting that CaM cannot regulate HNRPAB expression when IQCH is reduced. Thus, IQCH or CaM cannot regulate HNRPAB expression alone. Moreover, we deleted the IQ motif of IQCH, which is required for binding to CaM. The co-IP results showed that the interaction of IQCH and CaM was disrupted when deleting the IQ motif of IQCH, and the expression of HNRPAB was decreased. Therefore, we suggested that the interaction of IQCH and CaM might be required for IQCH regulating HNRPAB. In future studies, we will further investigate the relationships among IQCH, CaM, and HNRPAB.
Reviewer #3 (Public Review):
(1) More background details are needed regarding the proteins involved, in particular IQ proteins and calmodulin. The authors state that IQ proteins are not well-represented in the literature, but do not state how many IQ proteins are encoded in the genome. They also do not provide specifics regarding which calmodulins are involved, since there are at least 5 family members in mice and humans. This information could help provide more granular details about the mechanism to the reader and help place the findings in context.
Response: Thanks to reviewer’s suggestion. We have provided additional background information regarding IQ-containing protein family members in humans and mice, as well as other IQ-containing proteins implicated in male fertility, in the Introduction section. Furthermore, we have supplemented the Introduction with background information concerning the association between CaM and male infertility.
(2) The mouse fertility tests could be improved with more depth and rigor. There was no data regarding copulatory plug rate; data was unclear regarding how many WT females were used for the male breeding tests and how many litters were generated; the general methodology used for the breeding tests in the Methods section was not very explicitly or clearly described; the sample size of n=3 for the male breeding tests is rather small for that type of assay; and, given that ICHQ appears to be expressed in testicular interstitial cells (Fig. S10) and somewhat in other organs (Fig. S2), another important parameter of male fertility that should be addressed is reproductive hormone levels (e.g., LH, FSH, and testosterone). While normal epididymal size in Fig. S3 suggests that hormone (testosterone) levels are normal, epididymal size and/or weight were not rigorously quantified.
Response: Thanks to reviewer’s comment. We have provided the data regarding copulatory plug rate and the average number of litters for breeding tests in revised Figure 3—figure supplement 2. The methodology used for the breeding tests has been revised to be more detailed and explicit in the revised Method section. Moreover, we have increased the sample size for male breeding tests to n=6. We measured the serum levels of FSH, LH, and Testosterone in the WT (9.3±1.9 ng/ml, 0.93±0.15 ng/ml, and 0.2±0.03 ng/ml) and Iqch KO mice (12±2 ng/ml, 1.17±0.2 ng/ml, and 0.2±0.04 ng/ml). There was no significant difference observed in the serum levels of reproductive hormones between WT and Iqch KO mice; therefore, we did not include the data in the study. Furthermore, we have added quantitative data on epididymal size in the revised Figure 3—figure supplement 2.
(3) The Western blots in Figure 6 should be rigorously quantified from multiple independent experiments so that there is stronger evidence supporting claims based on those assays.
Response: We appreciate the reviewer's comment. As suggested, we have added quantified data in Figure 6—figure supplement 2 from the results of Western blotting in Figure 6.
(4) Some of the mouse testis images could be improved. For example, the PNA and PLCz images in Figure S7 are difficult to interpret in that the tubules do not appear to be stage-matched, and since the authors claimed that testicular histology is unaffected in knockout testes, it should be feasible to stage-match control and knockout samples. Also, the anti-ICHQ and CaM immunofluorescence in Figure S10 would benefit from some cell-type-specific co-stains to more rigorously define their expression patterns, and they should also be stage-matched.
Response: Thanks to reviewer’s suggestions. We have included immunofluorescence images of anti-PLCz, anti-PNA and anti-IQCH and CaM during spermatogenesis development.
Recommendations for the authors:
Reviewer #1 (Recommendations For The Authors):
(1) There are multiple grammatical errors and statements drawn beyond the results. The entire manuscript would benefit from professional editing.
Response: We are sorry for the grammatical errors. We have enlisted professional editing services to refine our manuscript.
(2) Line 40, "Firstly" is not appropriate here.
Response: Thanks to reviewer’s comment. The word "Firstly" has been removed from the revised manuscript.
(3) Line 44, "processes".
Response: Thanks to reviewer’s suggestion. We have changed “process” in to “processes” on line 45.
(4) "spermatocytogenesis (mitosis)" is incorrect.
Response: Thanks to reviewer’s comment. We have changed “spermatocytogenesis (mitosis)” in to “mitosis” on line 47.
(5) Ca and Ca2+ are both used in line 67 - 77. Be consistent.
Response: We appreciate the reviewer's detailed checks. We have maintained consistency by revising instances of "Ca" to "Ca2+" in revised manuscript.
(6) Line 238 to 240, "To elucidate the molecular mechanism by which IQCH regulates male fertility, we performed liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis using mouse sperm lysates and detected 288 interactors of IQCH (Data S1)."It is not clear how LC-MS/MS using mouse sperm lysates could detect "288 interactors of IQCH"? A co-IP experiment for IQCH using sperm lysates prior to LC-MS/MS is needed to detect "interactors of IQCH". However, in the Methods section, consistent with the main text, proteomic quantification was conducted for protein extract from sperm. Figure legend for Fig. 5 did not explain this, either.Thus, it is unable to evaluate Figure 5.
Response: We sincerely apologize for the oversight. Following reviewer’s suggestions, we have supplemented the method details of LC-MS/MS experiment in the Methods section of revised manuscript. Additionally, we conducted a co-IP experiment for IQCH using sperm lysates prior to LC-MS/MS and we did not include the corresponding figure in the manuscript. The results are as follows:
Author response image 1.
The results of a co-IP experiment for IQCH using sperm lysates from WT mice.
(7) Line 246, "... key proteins that might be activated by IQCH". What does "activated" here refer to? Should it be "upregulated"?
Response: We are sorry to our inexact statement. Instead, "upregulated" would better convey the intended meaning. According to reviewer’s suggestions, we have modified "activated" into "upregulated".
(8) Line 252 to 254, "the cross-analysis revealed that 76 proteins were shared between the IQCH-bound proteins and the IQCH-activated proteins (Fig. 5E), implicating this subset of genes as direct targets." This is a confusing statement. Is the author trying to say, IQCH-bound proteins have upregulated expression, suggesting that IQCH enhances their expression?
Response: We appreciate the reviewer's comment regarding the clarity of the statement in Line 252 to 254 of the manuscript. We have modified this sentence into “Importantly, cross-analysis revealed that 76 proteins were shared between the IQCH-bound proteins and the downregulated proteins in Iqch KO mice (Figure 5E), suggesting that IQCH might regulate their expression by the interaction.”
(9) Line 260 to 261, "SYNCRIP, HNRNPK, FUS, EWSR1, ANXA7, SLC25A4, and HNRPAB ... the loss of which showed the greatest influence on the phenotype of the Iqch KO mice." There is no evidence suggesting that the loss of SYNCRIP, HNRNPK, FUS, EWSR1, ANXA7, SLC25A4, and HNRPAB leads to Iqch KO phenotype.
Response: We apologize for our inaccurate statement. According to the literature, Fus KO, Ewsr1 KO, and Hnrnpk KO male mice were infertile, showing the spermatogenic arrest with absence of spermatozoa (Kuroda et al. 2000; Tian et al. 2021; Xu et al. 2022). Syncrip is involved meiotic process in Drosophila by interacting with Doublefault (Sechi et al. 2019). HNRPAB might be associated with mouse spermatogenesis by binding to Protamine 2 and contributing its translational regulation. Specifically, ANXA7 is a calcium-dependent phospholipid-binding protein that is a negative regulator of mitochondrial apoptosis (Du et al. 2015). Loss of SLC25A4 results in mitochondrial energy metabolism defects in mice (Graham et al. 1997). Moreover, RNA immunoprecipitation on formaldehyde cross-linked sperm followed by qPCR detected the interactions between HNRPAB and Catsper1, Catsper2, Catsper3, Ccdc40, Ccdc39, Ccdc65, Dnah8, Irrc6, and Dnhd1, which are essential for sperm development (Fukuda et al. 2013). Our Iqch KO mice showed abnormal sperm count, motility, morphology, and mitochondria, so we inferenced that IQCH might play a role in spermatogenesis by regulating the expression of SYNCRIP, HNRNPK, FUS, EWSR1, ANXA7, SLC25A4, and HNRPAB to some extent. We have changed an appropriate stamen that “We focused on SYNCRIP, HNRNPK, FUS, EWSR1, ANXA7, SLC25A4, and HNRPAB, which play important roles in spermatogenesis.”
(10) Fig. 6C and 6D use different styles of error bars.
Response: We are sorry for our oversight. In accordance with the reviewer's recommendations, we have modified the representation of error bars in the revised Fig. 6C.
(11) Line 296 to 297, "As expected, CaM interacted with IQCH, as indicated by LC-MS/MS analysis". It is not clear how LC-MS/MS detects protein interaction.
Response: As reviewer’s suggestions, we have supplemented the method details of LC-MS/MS experiment in the Methods section of revised manuscript. The results of proteins interacting with IQCH in sperm lysates from the LC-MS/MS experiment analysis were submitted as Figure 5—source data 1.
(12) It is still not clear how the interaction between IQCH, CaM, and HNRPAB is required for the expression of each other.
Response: Thank you for the reviewer’s comment. IQCH is a calmodulin-binding protein, and the binding of IQCH and CaM was confirmed by LC-MS/MS analysis and co-IP assay using sperm lysate. We thus speculated that if the interaction of IQCH and CaM might be a prerequisite for IQCH function. To prove that speculation, we took HNRPAB as an example. We knocked down IQCH in cultured cells, and a decrease in the expression of HNRPAB was observed. Similarly, when we knocked down CaM in cultured cells, and a decrease in the expression of HNRPAB was also detected. However, these results cannot exclude that IQCH or CaM could regulate HNRPAB expression alone. To investigate that if IQCH or CaM could regulate HNRPAB expression alone, we overexpressed IQCH in cells that knocked down CaM, while the expression of HNRPAB cannot be rescued, suggesting that IQCH cannot regulate HNRPAB expression when CaM is reduced. In consistent, we overexpressed CaM in cells that knocked down IQCH, while the expression of HNRPAB cannot be rescued, suggesting that CaM cannot regulate HNRPAB expression when IQCH is reduced. Thus, IQCH or CaM cannot regulate HNRPAB expression alone. Moreover, we deleted the IQ motif of IQCH, which is required for binding to CaM. The co-IP results showed that the interaction of IQCH and CaM was disrupted when deleting the IQ motif of IQCH, and the expression of HNRPAB was decreased. Therefore, we suggested that the interaction of IQCH and CaM might be required for IQCH regulating HNRPAB. In future studies, we will further investigate the relationships among IQCH, CaM, and HNRPAB.
Reviewer #3 (Recommendations For The Authors):
The authors have addressed my minor concerns. However, they neglected to address any of my more significant concerns in the public review. I assume that they simply overlooked these critiques, despite the fact that eLife explicitly states that "...as a general rule, concerns about a claim not being justified by the data should be explained in the public review." Therefore, the authors should have looked more carefully at the public reviews. As a result, my major concerns about the manuscript remain.
Response: We apologize for overlooking the public review process. We have improved our study based on the feedback received during the public review.
-

eLife assessment
This valuable study describes mice with a knock out of the IQ motif-containing H (IQCH) gene, to model a human loss-of-function mutation in IQCH associated with male sterility. The infertility is reproduced in the mouse, making it a compelling model, but some of the mechanistic experiments provide only indirect and thus incomplete evidence for interaction between IQCH and potential RNA binding proteins. With more rigorous approaches, the paper should be of interest to cell biologists and male reproductive biologists working on the sperm flagellar cytoskeleton and mitochondrial structure.
-

Reviewer #1 (Public Review):
By identifying a loss of function mutant of IQCH in infertile patient, Ruan et al. shows that IQCH is essential for spermiogenesis by generating a knockout mouse model of IQCH. Similar to infertile patient with mutant of IQCH, Iqch knockout mice are characterized by a cracked flagellar axoneme and abnormal mitochondrial structure. Mechanistically, IQCH regulates the expression of RNA-binding proteins (especially HNRPAB), which are indispensable for spermatogenesis.
Although this manuscript contains a potentially interesting piece of work that delineates a mechanism of IQCH that associates with spermatogenesis, this reviewer feels that a number of issues require clarification and re-evaluation for a better understanding of the role of IQCH in spermatogenesis.
Line 251 - 253, "To elucidate the molecular …
Reviewer #1 (Public Review):
By identifying a loss of function mutant of IQCH in infertile patient, Ruan et al. shows that IQCH is essential for spermiogenesis by generating a knockout mouse model of IQCH. Similar to infertile patient with mutant of IQCH, Iqch knockout mice are characterized by a cracked flagellar axoneme and abnormal mitochondrial structure. Mechanistically, IQCH regulates the expression of RNA-binding proteins (especially HNRPAB), which are indispensable for spermatogenesis.
Although this manuscript contains a potentially interesting piece of work that delineates a mechanism of IQCH that associates with spermatogenesis, this reviewer feels that a number of issues require clarification and re-evaluation for a better understanding of the role of IQCH in spermatogenesis.
Line 251 - 253, "To elucidate the molecular mechanism by which IQCH regulates male fertility, we performed liquid chromatography tandem mass spectrometry (LC‒MS/MS) analysis using mouse sperm lysates and detected 288 interactors of IQCH (Figure 5-source data 1)."
The reviewer had already raised significant concerns regarding the text above, noting that "LC‒MS/MS analysis using mouse sperm lysates" would not identify interactors of IQCH. However, this issue was not addressed in the revised manuscript. In the Methods section detailing LC-MS/MS, the authors stated that it was conducted on "eluates obtained from IP". However, there was no explanation provided on how IP for LC-MS/MS was performed. Additionally, it was unclear whether LC-MS or LC-MS/MS was utilized. The primary concern is that if LC‒MS/MS was conducted for the IP of IQCH, IQCH itself should have been detected in the results; however, as indicated by Figure 5-source data 1, IQCH was not listed.
-

Reviewer #3 (Public Review):
In this study, Ruan et al. investigate the role of the IQCH gene in spermatogenesis, focusing on its interaction with calmodulin and its regulation of RNA-binding proteins. The authors examined sperm from a male infertility patient with an inherited IQCH mutation as well as Iqch CRISPR knockout mice. The authors found that both human and mouse sperm exhibited structural and morphogenetic defects in multiple structures, leading to reduced fertility in Ichq-knockout male mice. Molecular analyses such as mass spectrometry and immunoprecipitation indicated that RNA-binding proteins are likely targets of IQCH, with the authors focusing on the RNA-binding protein HNRPAB as a critical regulator of testicular mRNAs. The authors used in vitro cell culture models to demonstrate an interaction between IQCH and …
Reviewer #3 (Public Review):
In this study, Ruan et al. investigate the role of the IQCH gene in spermatogenesis, focusing on its interaction with calmodulin and its regulation of RNA-binding proteins. The authors examined sperm from a male infertility patient with an inherited IQCH mutation as well as Iqch CRISPR knockout mice. The authors found that both human and mouse sperm exhibited structural and morphogenetic defects in multiple structures, leading to reduced fertility in Ichq-knockout male mice. Molecular analyses such as mass spectrometry and immunoprecipitation indicated that RNA-binding proteins are likely targets of IQCH, with the authors focusing on the RNA-binding protein HNRPAB as a critical regulator of testicular mRNAs. The authors used in vitro cell culture models to demonstrate an interaction between IQCH and calmodulin, in addition to showing that this interaction via the IQ motif of IQCH is required for IQCH's function in promoting HNRPAB expression. In sum, the authors concluded that IQCH promotes male fertility by binding to calmodulin and controlling HNRPAB expression to regulate the expression of essential mRNAs for spermatogenesis. These findings provide new insight into molecular mechanisms underlying spermatogenesis and how important factors for sperm morphogenesis and function are regulated.
The strengths of the study include the use of mouse and human samples, which demonstrate a likely relevance of the mouse model to humans; the use of multiple biochemical techniques to address the molecular mechanisms involved; the development of a new CRISPR mouse model; ample controls; and clearly displayed results. Assays are done rigorously and in a quantitative manner. Overall, the claims made by the authors in this manuscript are well-supported by the data provided.
-

-

Author Response
The following is the authors’ response to the original reviews.
Reviewer #1 (Recommendations For The Authors):
- Line 99-100 The authors claimed that IQCH is a novel IQ motif-containing protein, which is essential for spermiogenesis and fertilization. However, it is not clear if the currently published paper named an ancient testis-specific IQ motif containing H gene that regulates specific transcript isoform expression during spermatogenesis.
Response: Thanks to the reviewer’s comment. Yes, IQCH is the ancient testis-specific IQ motif containing H gene. According to the reviewer’s suggestion, we have revised the statement “Here, we revealed a testis-specific IQ motif containing H gene, IQCH, which is essential for spermiogenesis and fertilization” in Introduction part of revised manuscript.
- Line 154-159 …
Author Response
The following is the authors’ response to the original reviews.
Reviewer #1 (Recommendations For The Authors):
- Line 99-100 The authors claimed that IQCH is a novel IQ motif-containing protein, which is essential for spermiogenesis and fertilization. However, it is not clear if the currently published paper named an ancient testis-specific IQ motif containing H gene that regulates specific transcript isoform expression during spermatogenesis.
Response: Thanks to the reviewer’s comment. Yes, IQCH is the ancient testis-specific IQ motif containing H gene. According to the reviewer’s suggestion, we have revised the statement “Here, we revealed a testis-specific IQ motif containing H gene, IQCH, which is essential for spermiogenesis and fertilization” in Introduction part of revised manuscript.
- Line 154-159 Immunofluorescence staining for the marker of the acrosome (peanut agglutinin: PNA) as well as the mitochondrial marker (Transcription Factor A, Mitochondrial: TFAM) was performed to confirm the deficiency of the acrosomes and mitochondria in the proband's spermatozoa. It seems that the spermatozoa acrosomes and mitochondria were severely defective in the proband. The authors should indicate IQCH's role in mitochondrial and acrosome function and IQCH's role in mitochondrial and acrosome function these points by explaining how IQCH is related to mitochondrial and acrosome deficiency. In addition to staining, other functional analyses should be performed to strengthen the claim of acrosome and mitochondrial defects.
Response: We appreciate the reviewer's valuable suggestion. Indeed, in our study, the results of multiomics analysis on WT and Iqch KO testes, including LC-MS/MS analysis, proteomic analysis, and RNA-seq analysis, found a potential role of IQCH in mitochondrial and acrosome function. GO analysis of these analysis indicated a significant enrichment in mitochondrial and acrosomal functions, including acrosomal vesicle, acrosome assembly, vesicle fusion with Golgi apparatus, mitochondrion organization, mitochondrial matrix, and so on. Among the enriched molecules, in particular, HNRNPK mainly expresses at Golgi phase and Cap phase (Biggiogera et al. 1993). ANXA7 is a calcium-dependent phospholipid-binding protein that is a negative regulator of mitochondrial apoptosis (Du et al. 2015). Loss of SLC25A4 results in mitochondrial energy metabolism defects in mice (Graham et al. 1997). Furthermore, we confirmed that IQCH interacted with HNRNPK, ANXA7, and SLC25A4 through Co-IP, and exhibited downregulation in the sperm of the Iqch KO mice by immunofluorescence and western blotting. Moreover, IQCH can bind to HNRPAB, which could influence the mRNAs level of Catsper-family, such as Catsper1, Catsper2, and Catsper3, which are crucial for acrosome development (Jin ZR et al). In addition, we also detected HNRPAB binding to Dnhd1, which affects mitochondria development (Tan C et al). Therefore, in addition to staining, the other functional analyses also have provided the evidence of acrosome and mitochondrial defects caused by IQCH absence.
- Line 180-182 IQCH knockout mice were generated. It is not clear why Mut-IQCH mice were not generated to be consistent with the human sequencing data.
Response: Thanks for reviewer’s comments. To understand IQCH's impact on fecundity in mice, we employed CRISPR-Cas9 to generate mice encoding the orthologous variant of IQCH387+1_387+10del detected in humans. Regrettably, due to sequence complexity, the designed sgRNA's specificity and efficiency were low, hindering successful Iqch knock-in mouse construction. Considering IQCH387+1_387+10del results in absent expression, we pursued Iqch knockout mice to explore IQCH's role in spermatogenesis.
- Line 241.Figure 5A Gene Ontology (GO) analysis of the IQCH-bound proteins revealed a particular enrichment in fertilization, sperm axoneme assembly, mitochondrial organization, calcium channel, and RNA processing. But these GO functions are not shown in Figure 5A. The entire Figure 5 should be revised to enhance readability.
Response: We sincerely apologize for the oversight. These GO functions were indeed identified during the analysis of IQCH-bound proteins. Regrettably, we unintentionally omitted these GO functions when creating the plots. We have revised the plots in Figure 5 in revised manuscript to enhance readability.
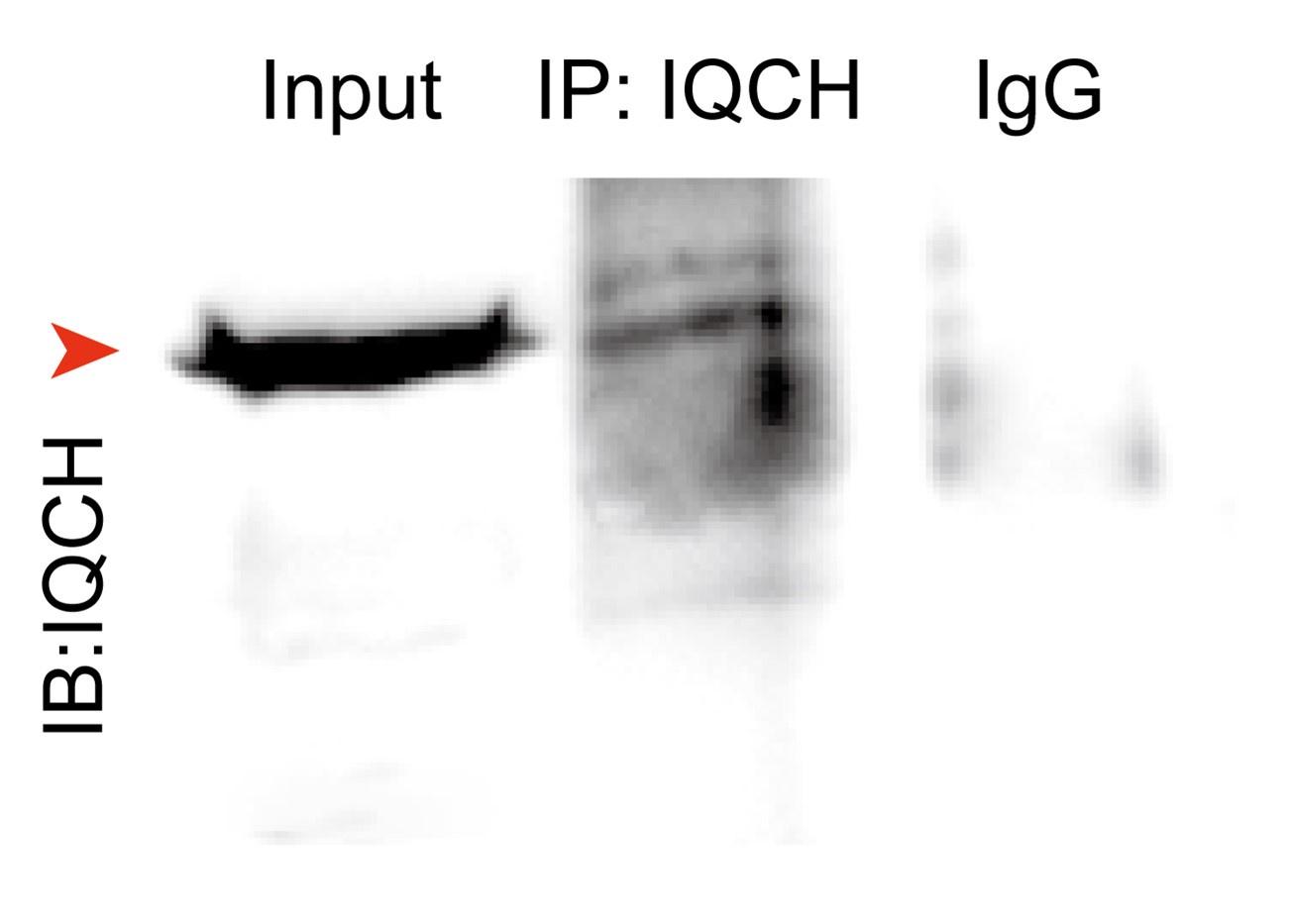
- Line 242 "33 ribosomal proteins were identified (Fig. 5B), indicating that IQCH might be involved in protein synthesis". The authors should perform an analysis to support the claim of protein synthesis defects.
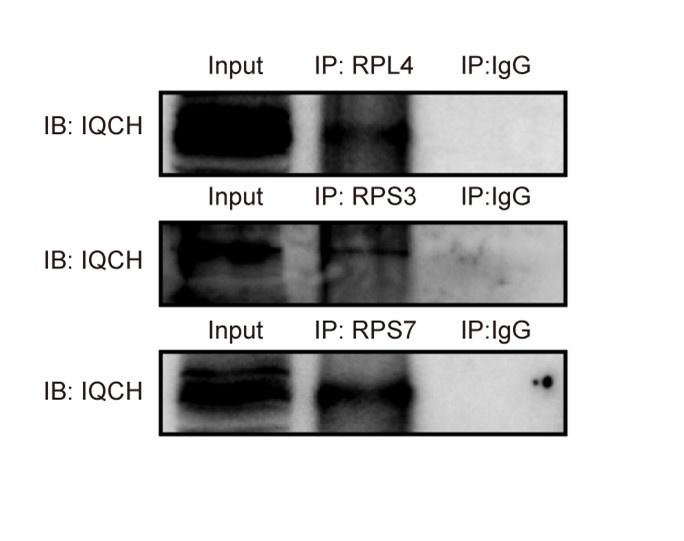
Response: Thanks to reviewer’s suggestions. Initially, we have supplemented Co-IP experiments to confirm the interaction between IQCH and three ribosomal proteins (RPL4, RPS3, and RPS7), chosen from a pool of 33 ribosomal proteins based on different protein scores (Figure R1). In addition, the proteomic analysis revealed 807 upregulated proteins and 1,186 downregulated proteins in KO mice compared to WT mice. We confirmed the key downregulated proteins by western blotting and immunofluorescence staining in the previous manuscript. These results indicated that IQCH might interact with ribosomal proteins to regulate protein expression. Naturally, the regulation of protein synthesis by IQCH requires further experiments for confirmation in future studies.
Author response image 1.
The interaction between IQCH and ribosomal proteins. Co-IP assays confirmed that IQCH interacted with RPL4, RPS3, and RPS7 in WT mouse sperm.
- Line 244 The authors mentioned too many GO functions without focus.
Response: Following reviewer’s suggestions, we have simplified IQCH-associated GO functions in the revised manuscript.
- Figure 6, there are no negative controls in all co-IP experiments. Band sizes are not marked. Thus, all data can't be evaluated. This also raises concern about whether the LC-MS/MS experiment to identify IQCH interacting protein was well-controlled? All co-IP experiments were poorly designed to draw any conclusion.
Response: Thanks to reviewer’s comments. We have supplemented negative controls in all Co-IP experiments and provided band sizes in Figure 6 in revised manuscript.
- The authors mentioned that IQCH can bind to CaM. But they didn't detect CaM protein in Figure 5. Did the LC-MS/MS experiment really work?
Response: Thanks to reviewer’s comments. We detected the interaction of CaM protein with IQCH in the LC-MS/MS experiment analysis, which has been submitted as new Data S1 in the revised manuscript. We also confirmed their binding in mouse sperm by Co-IP experiment and immunofluorescence staining, which results were shown in Figure 6 and Figure S10 in the previous study.
- Figure 6D. Because IQCH is lost in Iqch KO sperm, what is the point of showing in the Co-IP assay that CaM does not bind to IQCH in Iqch KO sperm?
Response: Following reviewer’s suggestions, we have deleted the results of Co-IP assay that CaM could not bind to IQCH in Iqch KO sperm.
- Figure 6E. The Co-IP assay does not support the authors' claim that the decreased expression of HNRPAB was due to the reduced binding of IQCH and CaM by the knockout of IQCH or CaM.
Response: Thanks to reviewer’s expert comments. Indeed, the results of Figure 6E confirmed the interaction of IQCH and CaM in K562 cells, and also showed that the expression of HNRPAB was reduced when IQCH or CaM was knocked down, suggesting that IQCH or CaM might regulate HNRPAB expression. While in Figure 6F, the downregulation of HNRPAB caused by knocking down IQCH (or CaM) cannot be rescued when overexpressed CaM (or IQCH), indicating that CaM (or IQCH) cannot mediate HNRPAB expression alone. Therefore, the reduced expression of HNRPAB in Figure 6E might result from the weakened interaction between IQCH and CaM, but not a superficial downregulation of IQCH or CaM expression. To avoid the confusion, we have modified the relevant description in the revied manuscript.
Reviewer #2 (Recommendations For The Authors):
Major comments:
- Lines 117 and 129: Please provide the reference number (NM_xxx.x) for the IQCH isoform that was used to interpret this variant. This is key information. Also, please provide the predicted truncation consequence caused by this splicing variant to IQCH protein.
Response: Thanks to reviewer’s suggestions. We have added reference number (NM_0010317152) of IQCH in manuscript. We employed splice site prediction tools, such as SpliceAI, RDDC, and varSEAK, to assess the expression consequences of this IQCH splicing variant. These tools couldn't anticipate the outcome of this splicing variant. However, the results of minigene splicing assay showed that the IQCH c.387+1_387+10del resulted in degradation of IQCH.
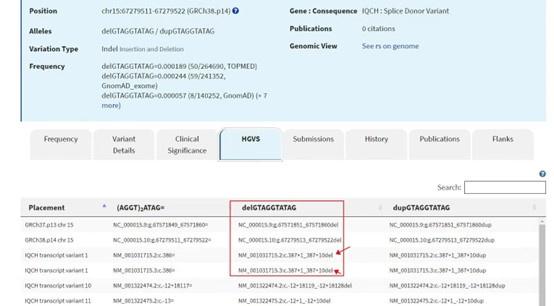
- Figure 1A: The deleted sequence indicated by the red box does not match IQCH c.387+1_387+10del. Please show a plot of the exon-intron boundary under the Sanger sequencing results of the WT allele.
Response: Thanks to reviewer’s suggestions. We are sorry for the use of non-standard descriptions about the results of Sanger sequencing. According to the HGVS nomenclature (Figure R2), we have modified the red box to match IQCH c.387+1_387+10del and have added the exon-intron boundary in Figure 1A accordingly.
Author response image 2.
HGVS nomenclature description of the IQCH variant. The picture showed a detailed HGVS nomenclature description of IQCH c.387+1_387+10del.
Minor comments:
a) Manuscript title: It is suggested to change the title to "IQCH regulates spermatogenesis by interacting with CaM to promote the expression of RNA-binding proteins".
Response: According to reviewer’s suggestions, we have modified the title as “IQCH regulates spermatogenesis by interacting with CaM to promote the expression of RNA-binding proteins”.
b) Line 116: Please introduce the abbreviation WES. Also, please introduce the other abbreviations (such as WT, SEM, TEM, etc.) the first time they appear.
Response: Thanks to reviewer’s suggestions. We have provided the full explanations for all abbreviations upon their initial appearance.
c) Line 140, "Nonfunctional IQCH": Due to "the lack of IQCH expression" in Line 137, should "Nonfunctional IQCH" be changed into "IQCH deficiency"?
Response: Thanks for reviewer’s the detailed review. We have modified this title in Results part of the revised manuscript as followed: “IQCH deficiency leads to sperm with cracked axoneme structures accompanied by defects in the acrosome and mitochondria”
d) The information on the following references is incomplete: Sechi et al., Tian et al., Wang et al., and Xu et al. Please provide issue/page/article numbers.
Response: We are sorry for our oversight. We have provided the missing issue/page/article numbers for the references.
e) The title of Figure 1: Please emphasize that the male infertile-associated variant is "homozygous".
Response: Thanks to reviewer’s suggestions. We have revised the title of Figure 1 to emphasize the homozygous variant as follows: “Identification of a homozygous splicing mutation in IQCH in a consanguineous family with male infertility”.
f) Table 1: Please provide the reference paper for the normal values. Response: We appreciate the reviewer's detailed checks. We have provided the reference paper for the normal values in Table 1.
g) Figure 5F is distorted. Please make sure that it is a perfect circle.
Response: Thanks to reviewer’s suggestions. We have revised both the graphical representation and layout of Figure 5 in revised manuscript to make sure the readability.
Reviewer #3 (Recommendations For The Authors):
While the writing is generally clear, there are multiple examples of where the writing could be improved for clarity.
- While some terms are defined throughout the manuscript, many abbreviations are not defined upon their first mention, such as WES, RT-PCR, TYH, HTF, KSOM, KEGG, RIPA, PMSE, SDS-PAGE, H&L, and HRP.
Response: Thanks to reviewer’s suggestions. We have provided the full explanations for all abbreviations upon their initial appearance.
- On line 44, the claim that spermatogenesis is the "most complex biological process" is rather subjective and hard to support with concrete data.
Response: Thanks to reviewer’s suggestions. We have modified this description in the Introduction section as follow: “Spermatogenesis is one of the most complex biological process in male organisms and functions to produce mature spermatozoa from spermatogonia in three phases: (i) spermatocytogenesis (mitosis), (ii) meiosis, and (iii) spermiogenesis.”
- On line 54, I think the authors meant "heterogeneous," not "heterologous."
Response: Thanks to reviewer’s comment. We have changed “heterologous” into “heterogeneous”.
- On line 156, I think the authors meant "deficiency," not "deficient."
Response: Thanks to reviewer’s comment. We are sorry to make this mistake. We have made the correction in the revised version of the manuscript.
- On line 300, K562 cells are mentioned, but neither in the Methods nor the Results are any details about the biological origin of these cells (or rationale for their use other than co-expression of IQCH and CaM) provided.
Response: Thanks to reviewer’s suggestion. K562 cell line is a human leukemia cell line and is enriched in the expression of IQCH and CaM, we thus opted to use this cell line for an easier knockdown of IQCH and CaM. We have supplemented the details about the biological origin of these cells in Method section of revised manuscript.
- For the Results section describing Figure 6H, it would be nice to provide some explanation of the results of ICHQ overexpression alone relative to control situations and not just relative to the delta-IQ version or relative to simultaneous CaM manipulation.
Response: According to the reviewer’s suggestion, we have supplemented the co-transfection of control and CaM plasmids in HEK293T cells, and the results showed that the expression of HNRPAB in cells co-transfected with control and CaM plasmids was similar to that of co-transfected with IQCH (△IQ) /CaM plasmids, but was lower than that in the cells overexpressing the WT-IQCH and CaM plasmids, confirming the nonfunction of IQCH (△IQ) plasmids. We have shown the results in Figure 6H in the revised manuscript.
- The sentence on lines 352-354 is confusing.
Response: We apologize for any confusion caused by the sentence in question. We have revisited the sentence and made appropriate revisions to enhance its clarity as follows: “Our findings suggest that the fertilization function is the main action of IQ motif-containing proteins, while each specific IQ motif-containing protein also has its own distinct role in spermatogenesis.”
- The use of "employee" on line 371 is awkward and not very scientific.
Response: Thanks to reviewer’s comment. We have changed “employee” in to “downstream effector protein” on line 376
-

eLife assessment
This valuable study describes mice with a knock out of the IQ motif-containing H (IQCH) gene, to model a human loss-of-function mutation in IQCH associated with male sterility. The infertility is reproduced in the mouse, making it a compelling model, but some of the mechanistic experiments provide only indirect and thus incomplete evidence for interaction between IQCH and potential RNA binding proteins. With more rigorous approaches, the paper should be of interest to cell biologists and male reproductive biologists working on the sperm flagellar cytoskeleton and mitochondrial structure.
-

Reviewer #1 (Public Review):
By identifying a loss of function mutant of IQCH in infertile patient, Ruan et al. shows that IQCH is essential for spermiogenesis by generating a knockout mouse model of IQCH. Similar to infertile patient with mutant of IQCH, Iqch knockout mice are characterized by a cracked flagellar axoneme and abnormal mitochondrial structure. Mechanistically, IQCH regulates the expression of RNA-binding proteins (especially HNRPAB), which are indispensable for spermatogenesis.
Although this manuscript contains a potentially interesting piece of work that delineates a mechanism of IQCH that associates with spermatogenesis, this reviewer feels that a number of issues require clarification and re-evaluation for a better understanding of the role of IQCH in spermatogenesis. With the shortage of logics and supporting data, …
Reviewer #1 (Public Review):
By identifying a loss of function mutant of IQCH in infertile patient, Ruan et al. shows that IQCH is essential for spermiogenesis by generating a knockout mouse model of IQCH. Similar to infertile patient with mutant of IQCH, Iqch knockout mice are characterized by a cracked flagellar axoneme and abnormal mitochondrial structure. Mechanistically, IQCH regulates the expression of RNA-binding proteins (especially HNRPAB), which are indispensable for spermatogenesis.
Although this manuscript contains a potentially interesting piece of work that delineates a mechanism of IQCH that associates with spermatogenesis, this reviewer feels that a number of issues require clarification and re-evaluation for a better understanding of the role of IQCH in spermatogenesis. With the shortage of logics and supporting data, causal relationships are still not clear among IQCH, CaM, and HNRPAB. The most serious point in this manuscript could be that the authors try to generalize their interpretations with too simplified model from limited pieces of their data. The way the data and the logic are presented needs to be largely revised, and several interpretations should be supported by direct evidence.
-

Reviewer #2 (Public Review):
The manuscript of "IQCH regulates spermatogenesis by interacting with CaM to promote RNA-binding proteins' expression" from Ruan et al. identified a homozygous variant affect the splicing of IQCH in two infertile men from a Chinese family. The authors also generated a Iqch knockout mouse model to confirm the abnormal sperm phenotypes associated with IQCH deficiency. Further molecular biological assays supported the important role and mechanism of IQCH in spermatogenesis. This manuscript is informative for the clinical and basic research of male infertility.
-

Reviewer #3 (Public Review):
In this study, Ruan et al. investigate the role of the IQCH gene in spermatogenesis, focusing on its interaction with calmodulin and its regulation of RNA-binding proteins. The authors examined sperm from a male infertility patient with an inherited IQCH mutation as well as Iqch CRISPR knockout mice. The authors found that both human and mouse sperm exhibited structural and morphogenetic defects in multiple structures, leading to reduced fertility in Ichq-knockout male mice. Molecular analyses such as mass spectrometry and immunoprecipitation indicated that RNA-binding proteins are likely targets of IQCH, with the authors focusing on the RNA-binding protein HNRPAB as a critical regulator of testicular mRNAs. The authors used in vitro cell culture models to demonstrate an interaction between IQCH and …
Reviewer #3 (Public Review):
In this study, Ruan et al. investigate the role of the IQCH gene in spermatogenesis, focusing on its interaction with calmodulin and its regulation of RNA-binding proteins. The authors examined sperm from a male infertility patient with an inherited IQCH mutation as well as Iqch CRISPR knockout mice. The authors found that both human and mouse sperm exhibited structural and morphogenetic defects in multiple structures, leading to reduced fertility in Ichq-knockout male mice. Molecular analyses such as mass spectrometry and immunoprecipitation indicated that RNA-binding proteins are likely targets of IQCH, with the authors focusing on the RNA-binding protein HNRPAB as a critical regulator of testicular mRNAs. The authors used in vitro cell culture models to demonstrate an interaction between IQCH and calmodulin, in addition to showing that this interaction via the IQ motif of IQCH is required for IQCH's function in promoting HNRPAB expression. In sum, the authors concluded that IQCH promotes male fertility by binding to calmodulin and controlling HNRPAB expression to regulate the expression of essential mRNAs for spermatogenesis. These findings provide new insight into molecular mechanisms underlying spermatogenesis and how important factors for sperm morphogenesis and function are regulated.
The strengths of the study include the use of mouse and human samples, which demonstrate a likely relevance of the mouse model to humans; the use of multiple biochemical techniques to address the molecular mechanisms involved; the development of a new CRISPR mouse model; ample controls; and clearly displayed results. There are some minor weaknesses in that more background details could be provided to the reader regarding the proteins involved; some assays could benefit from more rigorous quantification; some of the mouse testis images and analyses could be improved; and larger sample sizes, especially for the male mouse breeding tests, could be increased. Overall, the claims made the authors in this manuscript are well-supported by the data provided, but there some technical issues that, if addressed, could increase the robustness and rigor of the study.
1. More background details are needed regarding the proteins involved, in particular IQ proteins and calmodulin. The authors state that IQ proteins are not well-represented in the literature, but do not state how many IQ proteins are encoded in the genome. They also do not provide specifics regarding which calmodulins are involved, since there are at least 5 family members in mice and humans. This information could help provide more granular details about the mechanism to the reader and help place the findings in context.
2. The mouse fertility tests could be improved with more depth and rigor. There was no data regarding copulatory plug rate; data was unclear regarding how many WT females were used for the male breeding tests and how many litters were generated; the general methodology used for the breeding tests in the Methods section was not very explicitly or clearly described; the sample size of n=3 for the male breeding tests is rather small for that type of assay; and, given that ICHQ appears to be expressed in testicular interstitial cells (Fig. S10) and somewhat in other organs (Fig. S2), another important parameter of male fertility that should be addressed is reproductive hormone levels (e.g., LH, FSH, and testosterone). While normal epididymal size in Fig. S3 suggests that hormone (testosterone) levels are normal, epididymal size and/or weight were not rigorously quantified.
3. The Western blots in Figure 6 should be rigorously quantified from multiple independent experiments so that there is stronger evidence supporting claims based on those assays.
4. Some of the mouse testis images could be improved. For example, the PNA and PLCz images in Figure S7 are difficult to interpret in that the tubules do not appear to be stage-matched, and since the authors claimed that testicular histology is unaffected in knockout testes, it should be feasible to stage-match control and knockout samples. Also, the anti-ICHQ and CaM immunofluorescence in Figure S10 would benefit from some cell-type-specific co-stains to more rigorously define their expression patterns, and they should also be stage-matched.
-

-

eLife assessment
This important study combines experiments on human mutation and making a mouse model lacking IQCH and the functional consequences on spermatogenesis. The mouse model is compelling but some of the analysis is indirect and incomplete and would benefit from more rigorous direct approaches. With the experimental evidence that supports direct interaction between IQCH and potential RNA binding proteins strengthened, this paper would be of interest to cell biologists and male reproductive biologists working on the sperm flagellar cytoskeleton and mitochondrial structure.
-

Reviewer #1 (Public Review):
By identifying a loss of function mutant of IQCH in an infertile patient, Ruan et al. show that IQCH is essential for spermiogenesis by generating a knockout mouse model of IQCH. Similar to infertile patients with a mutant of IQCH, IQCH knockout mice are characterized by a cracked flagellar axoneme and abnormal mitochondrial structure. Mechanistically, IQCH regulates the expression of RNA-binding proteins (especially HNRPAB), which are indispensable for spermatogenesis.
Although this manuscript contains a potentially interesting piece of work that delineates a mechanism of IQCH that associates with spermatogenesis, this reviewer feels that a number of issues require clarification and re-evaluation for a better understanding of the role of IQCH in spermatogenesis. With the shortage of logic and supporting …
Reviewer #1 (Public Review):
By identifying a loss of function mutant of IQCH in an infertile patient, Ruan et al. show that IQCH is essential for spermiogenesis by generating a knockout mouse model of IQCH. Similar to infertile patients with a mutant of IQCH, IQCH knockout mice are characterized by a cracked flagellar axoneme and abnormal mitochondrial structure. Mechanistically, IQCH regulates the expression of RNA-binding proteins (especially HNRPAB), which are indispensable for spermatogenesis.
Although this manuscript contains a potentially interesting piece of work that delineates a mechanism of IQCH that associates with spermatogenesis, this reviewer feels that a number of issues require clarification and re-evaluation for a better understanding of the role of IQCH in spermatogenesis. With the shortage of logic and supporting data, causal relationships are still not clear among IQCH, CaM, and HNRPAB. The most serious point in this manuscript could be that the authors try to generalize their interpretations with a model that is too simplified from limited pieces of their data. The way the data and the logic are presented needs to be largely revised, and several interpretations should be supported by direct evidence.
-

Reviewer #2 (Public Review):
The manuscript "IQCH regulates spermatogenesis by interacting with CaM to promote RNA-binding proteins' expression" by Ruan et al. identified a homozygous variant affecting the splicing of IQCH in two infertile men from a Chinese family. The authors also generated an IQCH knockout mouse model to confirm the abnormal sperm phenotypes associated with IQCH deficiency. Further molecular biological assays supported the important role and mechanism of IQCH in spermatogenesis. This manuscript is informative for clinical and basic research on male infertility.
-

Reviewer #3 (Public Review):
In this study, Ruan et al. investigate the role of the IQCH gene in spermatogenesis, focusing on its interaction with calmodulin and its regulation of RNA-binding proteins. The authors examined sperm from a male infertility patient with an inherited IQCH mutation as well as IQCH CRISPR knockout mice. The authors found that both human and mouse sperm exhibited structural and morphogenetic defects in multiple structures, leading to reduced fertility in ICHQ-knockout male mice. Molecular analyses such as mass spectrometry and immunoprecipitation indicated that RNA-binding proteins are likely targets of IQCH, with the authors focusing on the RNA-binding protein HNRPAB as a critical regulator of testicular mRNAs. The authors used in vitro cell culture models to demonstrate an interaction between IQCH and …
Reviewer #3 (Public Review):
In this study, Ruan et al. investigate the role of the IQCH gene in spermatogenesis, focusing on its interaction with calmodulin and its regulation of RNA-binding proteins. The authors examined sperm from a male infertility patient with an inherited IQCH mutation as well as IQCH CRISPR knockout mice. The authors found that both human and mouse sperm exhibited structural and morphogenetic defects in multiple structures, leading to reduced fertility in ICHQ-knockout male mice. Molecular analyses such as mass spectrometry and immunoprecipitation indicated that RNA-binding proteins are likely targets of IQCH, with the authors focusing on the RNA-binding protein HNRPAB as a critical regulator of testicular mRNAs. The authors used in vitro cell culture models to demonstrate an interaction between IQCH and calmodulin, in addition to showing that this interaction via the IQ motif of IQCH is required for IQCH's function in promoting HNRPAB expression. In sum, the authors concluded that IQCH promotes male fertility by binding to calmodulin and controlling HNRPAB expression to regulate the expression of essential mRNAs for spermatogenesis. These findings provide new insight into molecular mechanisms underlying spermatogenesis and how important factors for sperm morphogenesis and function are regulated.
The strengths of the study include the use of mouse and human samples, which demonstrate a likely relevance of the mouse model to humans; the use of multiple biochemical techniques to address the molecular mechanisms involved; the development of a new CRISPR mouse model; ample controls; and clearly displayed results. There are some minor weaknesses in that more background details could be provided to the reader regarding the proteins involved; some assays could benefit from more rigorous quantification; some of the mouse testis images and analyses could be improved; and larger sample sizes, especially for the male mouse breeding tests, could be increased. Overall, the claims made by the authors in this manuscript are well-supported by the data provided and there are only minor technical issues that could increase the robustness and rigor of the study.
1. More background details are needed regarding the proteins involved, in particular IQ proteins and calmodulin. The authors state that IQ proteins are not well-represented in the literature, but do not state how many IQ proteins are encoded in the genome. They also do not provide specifics regarding which calmodulins are involved, since there are at least 5 family members in mice and humans. This information could help provide more granular details about the mechanism to the reader and help place the findings in context.
2. The mouse fertility tests could be improved with more depth and rigor. There was no data regarding copulatory plug rate; data was unclear regarding how many WT females were used for the male breeding tests and how many litters were generated; the general methodology used for the breeding tests in the Methods section was not very explicitly or clearly described; the sample size of n=3 for the male breeding tests is rather small for that type of assay; and, given that ICHQ appears to be expressed in testicular interstitial cells (Fig. S10) and somewhat in other organs (Fig. S2), another important parameter of male fertility that should be addressed is reproductive hormone levels (e.g., LH, FSH, and testosterone).
3. The Western blots in Figure 6 should be rigorously quantified from multiple independent experiments so that there is stronger evidence supporting claims based on those assays.
4. Some of the mouse testis images could be improved. For example, the PNA and PLCz images in Figure S7 are difficult to interpret in that the tubules do not appear to be stage-matched, and since the authors claimed that testicular histology is unaffected in knockout testes, it should be feasible to stage-match control and knockout samples. Also, the anti-ICHQ and CaM immunofluorescence in Figure S10 would benefit from some cell-type-specific co-stains to more rigorously define their expression patterns, and they should also be stage-matched.
-