Associations of four biological age markers with child development: A multi-omic analysis in the European HELIX cohort
Curation statements for this article:-
Curated by eLife
eLife assessment
This is an important study that examined multiple biological age measures in children, which has been lacking in literature. The findings of this study provided convincing evidence to interpret and understand the aging and developmental processes in children.
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
While biological age in adults is often understood as representing general health and resilience, the conceptual interpretation of accelerated biological age in children and its relationship to development remains unclear. We aimed to clarify the relationship of accelerated biological age, assessed through two established biological age indicators, telomere length and DNA methylation age, and two novel candidate biological age indicators, to child developmental outcomes, including growth and adiposity, cognition, behavior, lung function and the onset of puberty, among European school-age children participating in the HELIX exposome cohort.
Methods:
The study population included up to 1173 children, aged between 5 and 12 years, from study centres in the UK, France, Spain, Norway, Lithuania, and Greece. Telomere length was measured through qPCR, blood DNA methylation, and gene expression was measured using microarray, and proteins and metabolites were measured by a range of targeted assays. DNA methylation age was assessed using Horvath’s skin and blood clock, while novel blood transcriptome and ‘immunometabolic’ (based on plasma proteins and urinary and serum metabolites) clocks were derived and tested in a subset of children assessed six months after the main follow-up visit. Associations between biological age indicators with child developmental measures as well as health risk factors were estimated using linear regression, adjusted for chronological age, sex, ethnicity, and study centre. The clock derived markers were expressed as Δ age (i.e. predicted minus chronological age).
Results:
Transcriptome and immunometabolic clocks predicted chronological age well in the test set ( r =0.93 and r =0.84 respectively). Generally, weak correlations were observed, after adjustment for chronological age, between the biological age indicators.
Among associations with health risk factors, higher birthweight was associated with greater immunometabolic Δ age, smoke exposure with greater DNA methylation Δ age, and high family affluence with longer telomere length.
Among associations with child developmental measures, all biological age markers were associated with greater BMI and fat mass, and all markers except telomere length were associated with greater height, at least at nominal significance (p<0.05). Immunometabolic Δ age was associated with better working memory (p=4 e–3) and reduced inattentiveness (p=4 e–4), while DNA methylation Δ age was associated with greater inattentiveness (p=0.03) and poorer externalizing behaviors (p=0.01). Shorter telomere length was also associated with poorer externalizing behaviors (p=0.03).
Conclusions:
In children, as in adults, biological aging appears to be a multi-faceted process and adiposity is an important correlate of accelerated biological aging. Patterns of associations suggested that accelerated immunometabolic age may be beneficial for some aspects of child development while accelerated DNA methylation age and telomere attrition may reflect early detrimental aspects of biological aging, apparent even in children.
Funding:
UK Research and Innovation (MR/S03532X/1); European Commission (grant agreement numbers: 308333; 874583).
Article activity feed
-

Author Response
Reviewer #1 (Public Review):
This study provided evidence to interpret and understand the aging and developmental processes in children. The main strength of the study is it measures a set of biological age measures and a set of developmental measures, thus providing multi-faceted evidence to explain the associations between aging and development in children. The main weakness of this study is that how to measure and test the aging hypothesis of "a buildup of biological capital model" and "wear and tear" is not well-explained. Why the observed associations between biological age measures and developmental measures could support the aforementioned aging theories?
Thank you. On reflection we agree that how to test the aging hypotheses of "a buildup of biological capital model" and "wear and tear" is not well-explained …
Author Response
Reviewer #1 (Public Review):
This study provided evidence to interpret and understand the aging and developmental processes in children. The main strength of the study is it measures a set of biological age measures and a set of developmental measures, thus providing multi-faceted evidence to explain the associations between aging and development in children. The main weakness of this study is that how to measure and test the aging hypothesis of "a buildup of biological capital model" and "wear and tear" is not well-explained. Why the observed associations between biological age measures and developmental measures could support the aforementioned aging theories?
Thank you. On reflection we agree that how to test the aging hypotheses of "a buildup of biological capital model" and "wear and tear" is not well-explained in the manuscript. We have addressed this issue in the point-by-point responses below:
- Abstract - conclusion: The aging hypothesis of "a buildup of biological capital model" and "wear and tear" were mentioned in the conclusion without an explanation of these theories in the previous section. Readers who are not experts in the field may not understand the logic.
We have replaced these phrases in the abstract with the following interpretation, which we hope will be more readily understood:
“Patterns of associations suggested that accelerated immunometabolic age may be beneficial for some aspects of child development while accelerated DNA methylation age and telomere attrition may reflect early detrimental aspects of biological ageing, apparent even in children.”
- Result - Biological age marker performance: the correlation between transcriptome age and chronological age is very strong (r =0.94). I am afraid that very little age-independent information could be captured by the transcriptome age. Is it possible to down-regulate the age dependency of the transcriptome age in the training process?
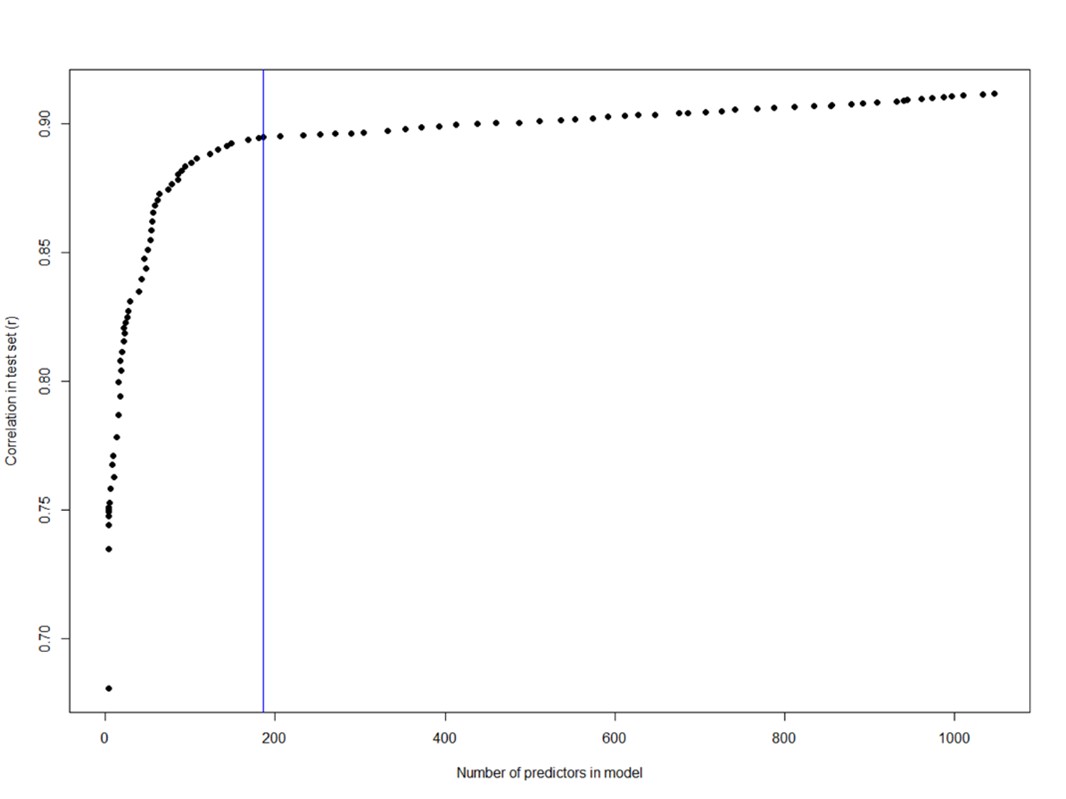
Thank you for this important comment: We agree the high accuracy of this clock may in fact reduce its relevance as a biological age marker and note that this is a concern generally in the field. We have explored the possibility of using a less accurate transcriptome age model as follows: Instead of elastic net modelling we tested using the lasso penalisation only, which will result in more parsimonious (sparse) models as less important features are dropped as the strength of the lambda parameter is increased. Plotting the correlation in the test set against number of features in models, as the lambda is sequentially increased, we can see (as shown in Author response image 1 by the blue line) that after the inclusion of around 200 features, the gain in accuracy becomes less steep.
Author response image 1.
We then tested the sensitivity of a model optimised for sparsity at the expense of some prediction accuracy, selected based on visual inspection (blue line, r in test set =0.87, number of features= 187) of the above plot, against developmental measures, compared to the most accurate model as presently included in the manuscript:
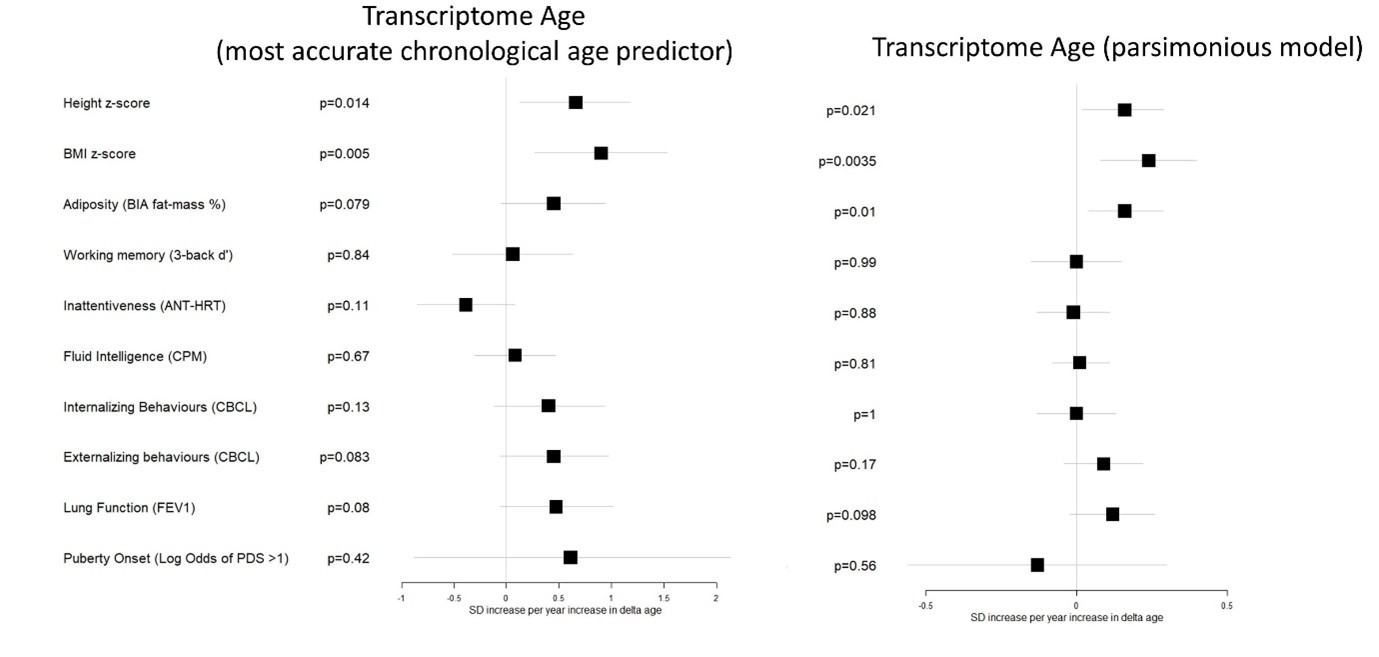
Author response image 2.
We find that, across all outcomes tested, the less accurate model, based on only the most important features, does not provide an improvement in sensitivity to developmental outcomes compared to the currently used model.
We therefore prefer to keep the more accurate model in this study. Especially as it is consistent with the methodology used in the Horvath and Immunometabolic age models and generally in the field, and otherwise it is not obvious how the biological clock should be trained (especially for children without mortality data) without altering the whole approach of the study. We have acknowledged and discussed this issue on page 15.
- The study population comes from several cohorts, which might influence the results. How the cohort effects were controlled for in the analyses?
The possible influence of cohort is a limitation of the study which we have discussed on page 16. We did not include cohort as a predictor in any of the candidate biological clocks since this may reduce detection of some age -related features. Instead, we include a variable for cohort as a fixed effect in all analyses with risk factors and developmental outcomes and examined the performance of candidate biological clocks in predicting chronological age within each cohort. As a further check, we have added an additional sensitivity analysis (Figure 4-figure supplement 6), against developmental outcomes significant in the main analysis, stratified by cohort. We find generally consistent effects across cohorts.
- Figure 3 only showed the number of p values. Can the author also provide the number of point estimates and 95% confidence intervals, perhaps in the supplemental table?
This information was originally provided in supplemental table 5 (now Supplementary file 7), combined with the sensitivity analyses. To make this information easier to find, we have made this a stand-alone table (table 3). We now direct readers to this information within the caption of Figure 4 (previously figure 2).
Reviewer #2 (Public Review):
The study had an especially relevant aim for aging research and utilized various data types in an especially interesting human population. Multi-omics perspective adds great value to the work. The researchers aimed to evaluate how different indicators of biological age (BA) behave in children during their developmental stage. In the analysis, relationships between indicators of BA, health risk factors, and developmental factors were assessed in cross-sectional data comprising children aged 5-12 years. The manuscript is well-written and easy to follow. The methodology is good. The authors succeeded to reach the aim in most parts.
In the study, previously known and unknown biological age indicators were used. Known indicators included telomere length and Horvath's epigenetic age. Unknown (novel) indicators, transcriptomic and immunometabolic clocks, were developed in the present study and they showed a strong correlation with calendar age in this population, also in the validation data set. Although the transcriptomic and immunometabolic clocks have the potential of being true indicators of biological age, they are still lacking scientific evidence of being such indicators in adults. That is, their associations with age-related diseases and mortality are yet to be shown. Thus, the major remark of the study relates to the phrasing: these novel transcriptomic and immunometabolic clocks should be presented as BA indicator candidates waiting for the needed evidence.
Thank you for this important observation. However, we still find that “biological age indicator” is a useful umbrella term in this manuscript and there is not an obvious alternative. We therefore have added the following sentence on page 8, and highlighted the difference between the markers at key points in the abstract, introduction, results and discussion.
“We note that since a common definition of markers of biological age is that they should be associated with age-related disease and mortality [69] these new clocks may only currently be considered “candidate” biological age markers. However, we have referred to both the established and candidate markers as biological age markers throughout to simplify presentation.”
-

-

eLife assessment
This is an important study that examined multiple biological age measures in children, which has been lacking in literature. The findings of this study provided convincing evidence to interpret and understand the aging and developmental processes in children.
-

Reviewer #1 (Public Review):
This study provided evidence to interpret and understand the aging and developmental processes in children. The main strength of the study is it measures a set of biological age measures and a set of developmental measures, thus providing multi-faceted evidence to explain the associations between aging and development in children. The main weakness of this study is that how to measure and test the aging hypothesis of "a buildup of biological capital model" and "wear and tear" is not well-explained. Why the observed associations between biological age measures and developmental measures could support the aforementioned aging theories?
1. Abstract - conclusion: The aging hypothesis of "a buildup of biological capital model" and "wear and tear" were mentioned in the conclusion without an explanation of these …
Reviewer #1 (Public Review):
This study provided evidence to interpret and understand the aging and developmental processes in children. The main strength of the study is it measures a set of biological age measures and a set of developmental measures, thus providing multi-faceted evidence to explain the associations between aging and development in children. The main weakness of this study is that how to measure and test the aging hypothesis of "a buildup of biological capital model" and "wear and tear" is not well-explained. Why the observed associations between biological age measures and developmental measures could support the aforementioned aging theories?
1. Abstract - conclusion: The aging hypothesis of "a buildup of biological capital model" and "wear and tear" were mentioned in the conclusion without an explanation of these theories in the previous section. Readers who are not experts in the field may not understand the logic.
2. Result - Biological age marker performance: the correlation between transcriptome age and chronological age is very strong (r =0.94). I am afraid that very little age-independent information could be captured by the transcriptome age. Is it possible to down-regulate the age dependency of the transcriptome age in the training process?
3. The study population comes from several cohorts, which might influence the results. How the cohort effects were controlled for in the analyses?
4. Figure 3 only showed the number of p values. Can the author also provide the number of point estimates and 95% confidence intervals, perhaps in the supplemental table? -

Reviewer #2 (Public Review):
The study had an especially relevant aim for aging research and utilized various data types in an especially interesting human population. Multi-omics perspective adds great value to the work. The researchers aimed to evaluate how different indicators of biological age (BA) behave in children during their developmental stage. In the analysis, relationships between indicators of BA, health risk factors, and developmental factors were assessed in cross-sectional data comprising children aged 5-12 years. The manuscript is well-written and easy to follow. The methodology is good. The authors succeeded to reach the aim in most parts.
In the study, previously known and unknown biological age indicators were used. Known indicators included telomere length and Horvath's epigenetic age. Unknown (novel) indicators, …
Reviewer #2 (Public Review):
The study had an especially relevant aim for aging research and utilized various data types in an especially interesting human population. Multi-omics perspective adds great value to the work. The researchers aimed to evaluate how different indicators of biological age (BA) behave in children during their developmental stage. In the analysis, relationships between indicators of BA, health risk factors, and developmental factors were assessed in cross-sectional data comprising children aged 5-12 years. The manuscript is well-written and easy to follow. The methodology is good. The authors succeeded to reach the aim in most parts.
In the study, previously known and unknown biological age indicators were used. Known indicators included telomere length and Horvath's epigenetic age. Unknown (novel) indicators, transcriptomic and immunometabolic clocks, were developed in the present study and they showed a strong correlation with calendar age in this population, also in the validation data set. Although the transcriptomic and immunometabolic clocks have the potential of being true indicators of biological age, they are still lacking scientific evidence of being such indicators in adults. That is, their associations with age-related diseases and mortality are yet to be shown. Thus, the major remark of the study relates to the phrasing: these novel transcriptomic and immunometabolic clocks should be presented as BA indicator candidates waiting for the needed evidence.
-