Two RNA-binding proteins mediate the sorting of miR223 from mitochondria into exosomes
Curation statements for this article:-
Curated by eLife
eLife assessment
This is an important study that reports the discovery of a new pathway of miRNA sorting to exosomes, involving a mitochondrially-localized protein. The evidence provided by some of the biochemical data is convincing. However, the major body of evidence is still incomplete.
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Fusion of multivesicular bodies (MVBs) with the plasma membrane results in the secretion of intraluminal vesicles (ILVs), or exosomes. The sorting of one exosomal cargo RNA, miR223, is facilitated by the RNA-binding protein, YBX1 (Shurtleff et al., 2016). We found that miR223 specifically binds a ‘cold shock’ domain (CSD) of YBX1 through a 5’ proximal sequence motif UCAGU that may represent a binding site or structural feature required for sorting. Prior to sorting into exosomes, most of the cytoplasmic miR223 resides in mitochondria. An RNA-binding protein localized to the mitochondrial matrix, YBAP1, appears to serve as a negative regulator of miR223 enrichment into exosomes. miR223 levels decreased in the mitochondria and increased in exosomes after loss of YBAP1. We observed YBX1 shuttle between mitochondria and endosomes in live cells. YBX1 also partitions into P body granules in the cytoplasm (Liu et al., 2021). We propose a model in which miR223 and likely other miRNAs are stored in mitochondria and are then mobilized by YBX1 to cytoplasmic phase condensate granules for capture into invaginations in the endosome that give rise to exosomes.
Article activity feed
-

-

Author Response
Reviewer #2 (Public Review):
The manuscript by Ma et al, "Two RNA-binding proteins mediate the sorting of miR223 from mitochondria into exosomes" examines the contribution of two RNA-binding proteins on the exosomal loading of miR223. The authors conclude that YBX1 and YBAP1 work in tandem to traffic and load miR223 into the exosome. The manuscript is interesting and potentially impactful. It proposes the following scenario regarding the exosomal loading of miR223: (1) YBAP1 sequesters miR223 in the mitochondria, (2) YBAP1 then transfers miR223 to YBX1, and (3) YBX1 then delivers miR223 into the early endosome for eventual secretion within an exosome. While the authors propose plausible explanations for this phenomenon, they do not specifically test them and no mechanism by which miR223 is shuttled between YBAP1 and …
Author Response
Reviewer #2 (Public Review):
The manuscript by Ma et al, "Two RNA-binding proteins mediate the sorting of miR223 from mitochondria into exosomes" examines the contribution of two RNA-binding proteins on the exosomal loading of miR223. The authors conclude that YBX1 and YBAP1 work in tandem to traffic and load miR223 into the exosome. The manuscript is interesting and potentially impactful. It proposes the following scenario regarding the exosomal loading of miR223: (1) YBAP1 sequesters miR223 in the mitochondria, (2) YBAP1 then transfers miR223 to YBX1, and (3) YBX1 then delivers miR223 into the early endosome for eventual secretion within an exosome. While the authors propose plausible explanations for this phenomenon, they do not specifically test them and no mechanism by which miR223 is shuttled between YBAP1 and YBX1, and the exosome is shown. Thus, the paper is missing critical mechanistic experiments that could have readily tested the speculative conclusions that it makes.
Comments:
- The major limitation of this paper is that it fails to explore the mechanism of any of the major changes it describes. For example, the authors propose that miR223 shuttles from mitochondrially localized YBAP1 to P-body-associated YBX1 to the exosome. This needs to be tested directly and could be easily addressed by showing a transfer of miR223 from YBAP1 to YBX1 to the exosome.
Testing this idea using fluorescently labeled miR223 would indeed be an ideal experiment. However, miRNA imaging presents challenges. As reviewer 1 pointed out, and we have now confirmed, the atto-647 dye itself localizes to mitochondria. We will continue our efforts to identify a suitable fluorescent label for miR223in order to be in a position to evaluate the temporal relationship between mitochondrial and endosomal miR223.
- If YBAP1 retains miR223 in mitochondria, what is the trigger for YBAP1 to release it and pass it off to YBX1? The authors speculate in their discussion that sequestration of mito-miR223 plays a "role in some structural or regulatory process, perhaps essential for mitochondrial homeostasis, controlled by the selective extraction of unwanted miRNA into RNA granules and further by secretion in exosomes...". This is readily testable by altering mitochondria dynamics and/or integrity.
A previous study has reported that YBAP1 can be released from mitochondria to the cytosol during HSV-1 infection (Song et al., 2021). However, due to restrictions, we are unable to conduct experiments using HSV to verify this condition. We attempted to induce mitochondrial stress by using different concentrations of CCCP, but we did not observe the release of YBAP1 from mitochondria after CCCP treatment. We speculate that not all mitochondrial stress conditions can trigger YBAP1 release. Investigating the mechanism of mito-miR223 release from mitochondria is one of our interests that we aim to explore in future studies.
- Much of the miRNA RT-PCR analysis is presented as a ratio of exosomal/cellular. This particular analysis assumes that cellular miRNA is unaffected by treatments. For example, Figure 1a shows that the presence of exosomal miR223 is significantly reduced when YBX1 is knocked out. This analysis does not consider the possibility that YBX1-KO alters (up or down-regulates) intracellular miR223 levels. Should that be the case, the ratiometric analysis is greatly skewed by intracellular miRNA changes. It would be better to not only show the intracellular levels of the miRs but also normalize the miRNA levels to the total amount of RNA isolated or an irrelevant/unchanged miRNA.
Our previous publications demonstrated that miR223 levels are increased in YBX1-KO cells and decreased in exosomes derived from YBX1 KO cells. However, no significant changes were observed in miR190 levels (Liu et al., 2021; Shurtleff et al., 2016). The repeated data has been included in Figure 1a.
For the analysis of other miRNAs by RT-PCR, we assessed changes in intracellular and exosomal miRNA levels in the corresponding figures. In the qPCR analysis, miRNA levels were normalized to the total amount of RNA.
- In figure 1, the authors show that in YBX1-KO cells, miR223 levels are decreased in the exosome. They further suggest this is because YBX1 binds with high affinity to miR223. This binding is compared to miR190 which the authors state is not enriched in the exosome. However, no data showing that miR190 is not present in the exosome is shown. A figure showing the amount of cellular and exosomal miR223 and 190 should be shown together on the same graph.
In previous publications we demonstrated that miR190 is not localized in exosomes and not significantly changed in YBX1 knockout (KO) cells and exosomes derived from YBX1 KO cells (Liu et al., 2021; Shurtleff et al., 2016). The repeated data has been included in Figure 1a.
- Figure 2 Supplement 1 - As to determine the nucleotides responsible for interacting with YBX1, the authors made several mutations within the miR223 sequence. However, no explanation is given regarding the mutant sequences used or what the ratios mean. Mutant sequences need to be included. How do the authors conclude that UCAGU is important when the locations of the mutations are unclear? Also, the interpretation of this data would benefit from a binding affinity curve as shown in Fig 2C.
The ratio is of labeled miR223/unlabeled miR223 (wt and mutant). All mutant sequences of miR223 have been included in Figure 2 supplement 1.
- While the binding of miR223mut to YBX1 is reduced, there is still significant binding. Does this mean that the 5nt binding motif is not exact? Do the authors know if there are multiple nucleotide possibilities at these positions that could facilitate binding? Perhaps confirming binding "in vivo" via RIP assay would further solidify the UCAGU motif as critical for binding to YBX1.
The binding affinity of miR223mut with YBX1 is reduced approximately 27-fold compared to miR223. We speculate that the secondary structure of miR223 may contribute to the interaction with YBX1.
Our EMSA data, in vitro packaging data, and exosome analysis reinforce the conclusion that UCAGU is critical for YBX1 binding. These findings suggest that the presence of the UCAGU motif in miR223 is crucial for its interaction with YBX1 and subsequent sorting into exosomes.
- Figures 2g, h - It would be nice to show that miR190mut also packages in the cell-free system. This would confirm that the sequence is responsible. Also, to confirm that the sorting of miR223 is YBX1-dependent, a cell-free reaction using cytosol and membranes from YBX1 KO cells is needed.
Although we have not performed the suggested experiment, we purified exosomes from cells overexpressing miR190sort and observed an increase in the enrichment of miR190sort in exosomes compared to miR190. This finding confirmed that the UCAGU motif facilitates miRNA sorting into exosomes.
Regarding the in vitro packaging assay, our previously published paper demonstrated that cytosol from YBX1 knockout (KO) cells significantly reduces the protection of miR223 from RNase digestion. We concluded that the sorting of miR223 into exosomes is dependent on YBX1 (Shurtleff et al., 2016).
- In Figure 3a, the authors show that miR223 is mitochondrially localized. Does the sequence of miR223 (WT or Mut) matter for localization? Does it matter for shuttling between YBAP1 and YBX1?
The localization of miR223mut has not been tested in our current study. We plan to conduct these experiments in the future.
- Supplement 3c - Is it strange that miR190 is not localized to any particular compartment? Is miR190 present ubiquitously and equally among all intracellular compartments?
Most mature miRNAs are predominantly localized in the cytoplasm. Although there is no specific subcellular localization reported for miR190 in the literature, our experimental findings indicate a relatively high expression of miR190 in 293T cells. It is likely that most of miR190 is localized in the cytosol. However, it is also possible that a small fraction of miR190 may associate with a membrane, which could explain its distribution in various subcellular structures. Importantly, we did not observe enrichment of miR190 in the mitochondria or exosomes.
- Figure 3h - Why would the miR223 levels increase if you remove mitochondria? Does CCCP also cause miR223 upregulation? I would have thought miR223 would just be mis-localized to the cytosol.
We report that the levels of cytoplasmic miR223 increase following the removal of mitochondria using CCCP treatment. While we cannot rule out the possibility that upregulation of miR223 is directly caused by CCCP treatment, we suggest that miR223 becomes mis-localized to the cytosol upon mitochondrial removal. Our data suggests that mitochondria contribute to the secretion of miR223 into exosomes. When mitochondria are removed by mitophagy, cytosolic miR223 is not efficiently secreted, which provides an alternative explanation for the observed increase in miR223 level after mitochondrial removal.
- Figure 3i - What is the meaning of "Urd" in the figure label? This isn't mentioned anywhere.
“Urd” represents Uridine. Uridine is now spelled out in figure 3i. The absence of mitochondria can impact the function of the mitochondrial enzyme dihydroorotate dehydrogenase, which plays a role in pyrimidine synthesis. To address this issue, one approach is to supplement the cell culture medium with Urd. A previous study demonstrated that primary fibroblasts showed positive responses when Urd was added to the cell culture medium, resulting in improved cell viability for extended periods of time (Correia-Melo et al., 2017).
- Figure 3j - The data is presented as a ratio of EV/cell. Again, this inaccurately represents the amount of miR223 in the EV. This issue is apparent when looking at Figures 3h and 3j. In 3h, CCCP causes an upregulation of intracellular miR223. As such, the presumed decrease in EV miR233 after CCCP (3j) could be an artifact due to increased levels of intracellular miR223. Both intracellular and EV levels of miRs need to be shown.
Both the intracellular and exosomal levels of miR223 have been included in Figure 3j.
- In Figure 4, the authors show that when overexpressed, YBX1 will pulldown YBAP1. Can the authors comment as to why none of the earlier purifications show this finding (Figure 1 for example)? Even more curious is that when YBAP1 is purified, YBX1 does not co-purify (Figure 4 supplement 1a, b).
In Figure 4a-b, human YBX1 fused with a Strep II tag was purified from 293T cells using Strep-Tactin® Sepharose® resin in a one-step purification process. Our data has shown that YBAP1 is expressed in 293T cells.
In Figure 1 and Figure 4 Supplement 1a, human YBX1 or YBAP1 fused with His and MBP tags were purified from insect cells using a three-step purification process involving Ni-NTA His-Pur resin, amylose resin, and Superdex-200 gel filtration chromatography.
One possibility is that human YBX1 or YBAP1 may not interact well with insect YBAP1 or YBX1, which could result in separate tagged forms of YBX1 or YBAP1 isolated from insect cells.
Another possibility is that the expression levels of insect YBAP1 and YBX1 may be too low. Consequently, tagged forms YBX1 or YBAP1 expressed in insect cells may copurify with partners not readily detected by Coomassie blue stain. However, in Figure 4 Supplement 1b, human YBX1 fused with His and MBP tags was co-expressed with non-tagged human YBAP1, and both bands of YBX1 and YBAP1 were visible on the Coomassie blue gel after purification using Ni-NTA His-Pur resin, amylose resin, and Superdex-200 gel filtration chromatography.
- Figure 4f, g - The text associated with these figures is very confusing, as is the labeling for the input. Also, what is "miR223 Fold change" in this regard? Seeing as your IgG should not have IP'd anything, normalizing to IgG can amplify noise. As such, RIP assays are typically presented as % input or fold enrichment.
The RIP assay results have been calculated and presented as a % input in Figure 4g.
- Figure 4h - The authors show binding between miR223 and YBAP1 however it is not clear how significant this binding is. There is more than a 30-fold difference in binding affinity between miR223 and YBX1 than between miR223 and YBAP1. Even more, when comparing the EMSAs and fraction bound from figures 1 and 2 to those of Figure 4h, the binding between miR223 and YBAP1 more closely resembles that of miR190 and YBX1, which the authors state is a non-binder of YBX1. The authors will need to reconcile these discrepancies.
We agree that the binding of YBAP and YBX1 differ quite significantly in the affinity of their interaction with miR223. It is difficult to draw conclusions from a comparison of the affinities of YBX1 for miR190 and YBAP1 for miR223. Nonetheless, a quantitative difference in the interaction of YBAP1 with miR223 and miR190 is apparent (Fig. 4 h, I, j) and we observed no enrichment miR190 in isolated mitochondria (Fig. 3 supplement 1a) whereas YBAP1 selectively IP’d miR223 from isolated mitochondria (Fig. 4 f and g).
- Can the authors present the Kd values for EMSA data?
The Kd values for the EMSA data have been added to the respective figures.
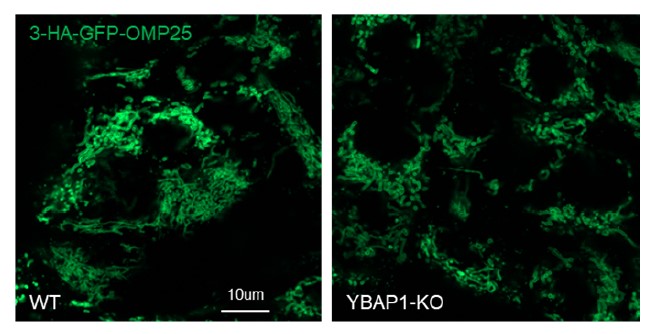
- Figure 5 - Does YBAP1-KO affect mitochondrial protein integrity or numbers?
We generated stable cell lines expressing 3xHA-GFP-OMP25 in both 293T WT and YBAP1-KO cells, but we did not observe any alterations in mitochondrial morphology (Author response image 1).
Author response image 1.
Additionally, we performed a comparison of different mitochondrial markers using immunoblot in 293T WT cells and YBAP1-KO cells and did not observe any changes in these markers (data has been included in Figure 5b.).
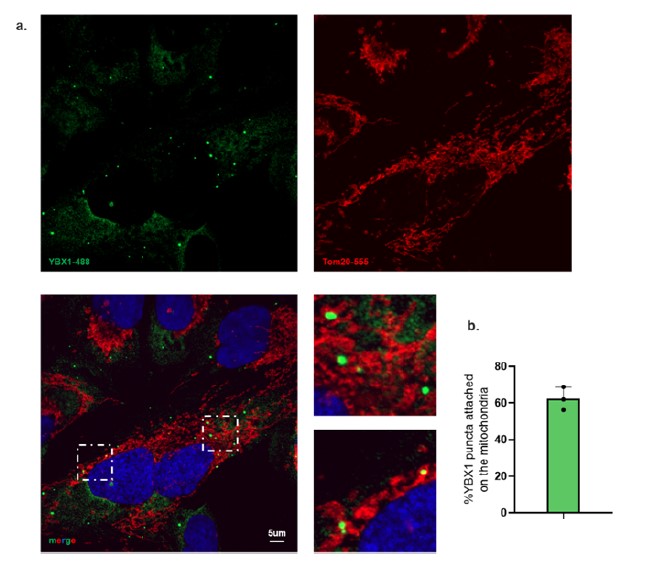
- Figure 6a - Are the authors using YBAP1 as their mitochondrial marker? Please include TOM20 and/or 22.
In Figure 4c and 4e, our data clearly demonstrate that the majority of YBAP1 is localized in the mitochondria.
To further validate this localization, we performed immunofluorescence staining using antibodies against endogenous Tom20 and YBX1. The immunofluorescence images document YBX1 associated with mitochondria (Author response image 2 and new Fig 6a.).
Author response image 2.
- Figure 6b - Rab5 is an early endosome marker and may not fully represent the organelles that become MVBs. Co-localization at this point does not suggest that associating proteins will be present in the exosome, and it is possible that the authors are looking at the precursor of a recycling endosome. Even more, exosome loading does not occur at the early endosome, but instead at the MVB. Perhaps looking at markers of the late endosome such as Rab7 or ideally markers of the MVB such as M6P or CD63 would help draw an association between YBX1, YBAP1, and the exosome. Also, If the authors want to make the claim that interactions at the early endosome leads to secretion as an exosome, the authors should show that isolated EVs from Rab5Q79L-expressing cells contain miR223.
We have previously used overexpressed Rab5(Q79L) to monitor the localization of exosomal content, specifically CD63 and YBX1, in enlarged endosomes (Liu et al. 2021, Fig. 4A, B). These endosomes exhibit a mixture of early and late endocytic markers, including CD63. (Wegner et al., 2010). Hence, the presence of Rab5(Q79L)-positive enlarged endosomes does not solely indicate early endosomes.
- The mentioning of P-bodies is interesting but at no time is an association addressed. This is therefore an overly speculative conclusion. Either show an association or leave this out of the manuscript.
In a previous paper we demonstrated that YBX1 puncta colocalize with P-body markers EDC4, Dcp1 and DDX6 (Liu et al., 2021).
- In lines 55-58, the authors make the comment "However, many of these studies used sedimentation at ~100,000 g to collect EVs, which may also collect RNP particles not enclosed within membranes which complicates the interpretation of these data." Do RNPs not dissolve when secreted? Can the authors give a reference for this statement?
In a previous paper, we demonstrated that the RNP Ago2 does not dissolve in the conditioned medium and is not in vesicles but sediments to the bottom of a density gradient (Temoche-Diaz et al., 2019).
-

eLife assessment
This is an important study that reports the discovery of a new pathway of miRNA sorting to exosomes, involving a mitochondrially-localized protein. The evidence provided by some of the biochemical data is convincing. However, the major body of evidence is still incomplete.
-

Reviewer #1 (Public Review):
This study focuses on molecular and cellular mechanisms underlying the sorting of miRNAs into exosomes originating from multivesicular bodies (MVBs). Following up on their previous work, the authors analysed the biochemical basis of miRNA selection by the RNA-binding protein YBX1 which is known to participate in this sorting. Using electrophoretic mobility shift assays (EMSA) involving a series of YBX1 constructs, they pinpointed the key role of the cold shock domain of YBX1 (supported by the C-terminal domain) in miRNA binding. By comparing a secreted model miRNA (miR223), a control cytoplasmic miRNA that is not enriched in exosomes (miR190), and a series of their swap mutants, the authors identified what could be a sequence motif enabling YBX1 to discriminate - through direct binding - between miRNAs to be …
Reviewer #1 (Public Review):
This study focuses on molecular and cellular mechanisms underlying the sorting of miRNAs into exosomes originating from multivesicular bodies (MVBs). Following up on their previous work, the authors analysed the biochemical basis of miRNA selection by the RNA-binding protein YBX1 which is known to participate in this sorting. Using electrophoretic mobility shift assays (EMSA) involving a series of YBX1 constructs, they pinpointed the key role of the cold shock domain of YBX1 (supported by the C-terminal domain) in miRNA binding. By comparing a secreted model miRNA (miR223), a control cytoplasmic miRNA that is not enriched in exosomes (miR190), and a series of their swap mutants, the authors identified what could be a sequence motif enabling YBX1 to discriminate - through direct binding - between miRNAs to be secreted or to be retained.
The authors then wondered from which subcellular pool miR223 could be mobilised for secretion. They turned their attention to the mitochondria and found evidence of miR223 association with these organelles. Interestingly, when mitochondria were depleted by Parkin overexpression and CCCP treatment, the cellular level of miR223, but not of miR190, increased, whereas its enrichment in extracellular vesicles dropped. This observation permitted to forward a hypothesis whereby mitochondria could be involved in miR223 mobilisation into exosomes. This process would be mediated by YBX1 which shuttles between mitochondria and endosomes, as was elegantly shown in live imaging experiments.
Finally, the authors provide initial data implicating in this process the mitochondrial matrix protein YBAP1, broadly known as C1QBP, or p32. YBAP1 was found to interact with YBX1 and miR223 in pull-down assays. Moreover, direct and moderately strong miR223 binding by YBAP1 was confirmed by EMSA. Interestingly, just like YBX1, YBAP1 seems to prefer this substrate over miR190, indicating certain binding specificity. The observation that YBAP1 knockout resulted in the decreased association of miR223 with mitochondria, paralleled by its correspondingly better mobilisation into exosomes, enabled the authors to propose that YBAP1 could negatively control miR223 secretion at the level of mitochondria.
Strengths
This is a very interesting study proposing an elegant hypothesis and featuring a creative panel of methods, many of which will certainly be of interest to biochemists and cell biologists working with extracellular RNA and mitochondria (e.g. the Parkin/CCCP-mediated mitochondria depletion and the time-lapse imaging of RNA-binding proteins against cellular organelles).
The authors did a good job of dissecting the YBX1 interaction with miR223 versus miR190. These experiments are performed at a high technical level, and their interpretation is straightforward and convincing. The nearly two orders of magnitude difference in affinity provides a plausible means by which YBX1 could recognise and funnel one, but not the other, miRNA into the secretion pathway.
Another valuable piece of data is related to YBAP1. This important, deeply conserved protein, strongly implicated in severe mitochondrial diseases and cancer, remains poorly understood at the level of basic molecular mechanisms, and even its subcellular localisation is debated. The data presented by the authors reinforce the idea of its primarily mitochondrial localisation, in agreement with earlier studies. They also provided new information about the RNA-binding activity of YBAP1. First proposed to interact with RNA by Yagi et al., Nucleic Acids Res 2012 (doi:10.1093/nar/gks774), YBAP1 is confirmed in the present study as a reasonably affine RNA-binding protein, based on direct EMSA experiments involving a highly purified protein and natural RNAs. These data should encourage the community to explore the full RNA-binding potential of YBAP1/C1QBP/p32 in a wider variety of models, especially in the context of mitochondrial gene expression.Weaknesses
While the authors might be right about the existence of a sequence motif that specifies miRNAs for exosome sorting by YBX1, it is at present difficult to disentangle the sequence and structure contributions to YBX1 binding within the variants described in the paper. RNA structure predictions, however imperfect, suggest that miR223-3p is a fully single-stranded transcript (ensemble ΔG = -0.33 kcal/mol, RNAfold), while miR190-5p is a tightly base-paired one (ensemble ΔG = -2.85 kcal/mol). This likely explains the differential affinity to YBX1, known to strongly prefer single-stranded RNAs. When mutating the putative sorting motif in miR223 (UCAGU>AGACA), the authors introduced some amount of secondary structure (ΔG = -1.04 kcal/mol), which could have impeded YBX1 binding. By contrast, the mutation of miR190 (AUAUG>UCAGU) significantly weakened the structure (ensemble ΔG = -2.21 kcal/mol), which might explain the improvement in YBX1 interaction.
Mitochondria appear to be a plausible location for mobilisable RNAs, given their multiple associations with ribosomes, RNA-containing condensates, and other organelles. However, the presented evidence of the mitochondrial localisation of miR223 is limited. The colocalisation pattern of the ATTO 647-labelled miR223 with the well-behaved mitochondrial marker Tom22 is remarkable; such a neat overlap has so far only been observed for some abundant mtDNA-encoded transcripts, but not for an extraneous transcript. The interpretation of this result will depend a great deal on experimental details which, unfortunately, are missing for this section. ATTO 647N is known to be quantitatively recruited to mitochondria, producing just the same kind of complete colocalisation, making it a perfect tool to visualise mitochondria in the cell (Han et al., Nat Commun 2017, doi:10.1038/s41467-017-01503-6). There is a worry that the colocalisation observed here might have been driven by the dye alone.
Furthermore, the definition of the topology of RNA localisation with respect to the mitochondrial membranes remains challenging, and a number of more robust methods have been recently proposed to address this contentious issue. At minima, one would expect that the authors would use RNase treatment, with or without Triton X-100 (like they did in the in vitro packaging assay), to see whether miR223 is indeed protected by the mitochondrial membranes and, therefore, resides in the interior of the organelles. As for now, based on the presented data, one can safely conclude that miR223 is associated with the mitochondria, without claiming that it is necessary inside them.
The Parkin/CCCP method is very powerful, which is its strength and weakness at the same time. miR223 secretion does decrease when the mitochondria are depleted. However, it is unclear how direct and specific this effect is. The destruction of mitochondria likely crashed the cellular ATP levels, which could have generally affected vesicular transport, not only miR223 sorting. A more detailed analysis of the overall abundance of extracellular vesicles and their cargo under these conditions could reveal the true scope of the mitochondrial contribution to RNA secretion.
YBAP1 is a difficult, indeed "treacherous", protein to work with. Its strong negative charge (pI = 4) makes it easily stick to positively charged proteins, such as YBX1 (pI = 9.9). Such interactions are routinely observed in pulldown assays from cell lysates, where all components are intermixed (but often cannot be corroborated by in situ or in vivo approaches). The authors carefully showed that YBX1 and YBAP1 do not significantly colocalise in the cell, which makes the interplay between the two proteins in miR223 sorting difficult to stage. They also studied the miR223 distribution between mitochondria and extracellular vesicles using YBAP1 knockout cells. However, such cells are known to be very sick and have an extremely pleiotropic mitochondrial and metabolic phenotype. Therefore, the apparent implication of YBAP1 in miR223 sorting might be less direct than currently envisaged.
-

Reviewer #2 (Public Review):
The manuscript by Ma et al, "Two RNA-binding proteins mediate the sorting of miR223 from mitochondria into exosomes" examines the contribution of two RNA-binding proteins on the exosomal loading of miR223. The authors conclude that YBX1 and YBAP1 work in tandem to traffic and load miR223 into the exosome. The manuscript is interesting and potentially impactful. It proposes the following scenario regarding the exosomal loading of miR223: (1) YBAP1 sequesters miR223 in the mitochondria, (2) YBAP1 then transfers miR223 to YBX1, and (3) YBX1 then delivers miR223 into the early endosome for eventual secretion within an exosome. While the authors propose plausible explanations for this phenomenon, they do not specifically test them and no mechanism by which miR223 is shuttled between YBAP1 and YBX1, and the …
Reviewer #2 (Public Review):
The manuscript by Ma et al, "Two RNA-binding proteins mediate the sorting of miR223 from mitochondria into exosomes" examines the contribution of two RNA-binding proteins on the exosomal loading of miR223. The authors conclude that YBX1 and YBAP1 work in tandem to traffic and load miR223 into the exosome. The manuscript is interesting and potentially impactful. It proposes the following scenario regarding the exosomal loading of miR223: (1) YBAP1 sequesters miR223 in the mitochondria, (2) YBAP1 then transfers miR223 to YBX1, and (3) YBX1 then delivers miR223 into the early endosome for eventual secretion within an exosome. While the authors propose plausible explanations for this phenomenon, they do not specifically test them and no mechanism by which miR223 is shuttled between YBAP1 and YBX1, and the exosome is shown. Thus, the paper is missing critical mechanistic experiments that could have readily tested the speculative conclusions that it makes.
Comments:
1. The major limitation of this paper is that it fails to explore the mechanism of any of the major changes it describes. For example, the authors propose that miR223 shuttles from mitochondrially localized YBAP1 to P-body-associated YBX1 to the exosome. This needs to be tested directly and could be easily addressed by showing a transfer of miR223 from YBAP1 to YBX1 to the exosome.
2. If YBAP1 retains miR223 in mitochondria, what is the trigger for YBAP1 to release it and pass it off to YBX1? The authors speculate in their discussion that sequestration of mito-miR223 plays a "role in some structural or regulatory process, perhaps essential for mitochondrial homeostasis, controlled by the selective extraction of unwanted miRNA into RNA granules and further by secretion in exosomes...". This is readily testable by altering mitochondria dynamics and/or integrity.
3. Much of the miRNA RT-PCR analysis is presented as a ratio of exosomal/cellular. This particular analysis assumes that cellular miRNA is unaffected by treatments. For example, Figure 1a shows that the presence of exosomal miR223 is significantly reduced when YBX1 is knocked out. This analysis does not consider the possibility that YBX1-KO alters (up or down-regulates) intracellular miR223 levels. Should that be the case, the ratiometric analysis is greatly skewed by intracellular miRNA changes. It would be better to not only show the intracellular levels of the miRs but also normalize the miRNA levels to the total amount of RNA isolated or an irrelevant/unchanged miRNA.
4. In figure 1, the authors show that in YBX1-KO cells, miR223 levels are decreased in the exosome. They further suggest this is because YBX1 binds with high affinity to miR223. This binding is compared to miR190 which the authors state is not enriched in the exosome. However, no data showing that miR190 is not present in the exosome is shown. A figure showing the amount of cellular and exosomal miR223 and 190 should be shown together on the same graph.
5. Figure 2 Supplement 1 - As to determine the nucleotides responsible for interacting with YBX1, the authors made several mutations within the miR223 sequence. However, no explanation is given regarding the mutant sequences used or what the ratios mean. Mutant sequences need to be included. How do the authors conclude that UCAGU is important when the locations of the mutations are unclear? Also, the interpretation of this data would benefit from a binding affinity curve as shown in Fig 2C.
6. While the binding of miR223mut to YBX1 is reduced, there is still significant binding. Does this mean that the 5nt binding motif is not exact? Do the authors know if there are multiple nucleotide possibilities at these positions that could facilitate binding? Perhaps confirming binding "in vivo" via RIP assay would further solidify the UCAGU motif as critical for binding to YBX1.
7. Figures 2g, h - It would be nice to show that miR190mut also packages in the cell-free system. This would confirm that the sequence is responsible. Also, to confirm that the sorting of miR223 is YBX1-dependent, a cell-free reaction using cytosol and membranes from YBX1 KO cells is needed.
8. In Figure 3a, the authors show that miR223 is mitochondrially localized. Does the sequence of miR223 (WT or Mut) matter for localization? Does it matter for shuttling between YBAP1 and YBX1?
9. Supplement 3c - Is it strange that miR190 is not localized to any particular compartment? Is miR190 present ubiquitously and equally among all intracellular compartments?
10. Figure 3h - Why would the miR223 levels increase if you remove mitochondria? Does CCCP also cause miR223 upregulation? I would have thought miR223 would just be mis-localized to the cytosol.
11. Figure 3i - What is the meaning of "Urd" in the figure label? This isn't mentioned anywhere.
12. Figure 3j - The data is presented as a ratio of EV/cell. Again, this inaccurately represents the amount of miR223 in the EV. This issue is apparent when looking at Figures 3h and 3j. In 3h, CCCP causes an upregulation of intracellular miR223. As such, the presumed decrease in EV miR233 after CCCP (3j) could be an artifact due to increased levels of intracellular miR223. Both intracellular and EV levels of miRs need to be shown.
13. In Figure 4, the authors show that when overexpressed, YBX1 will pulldown YBAP1. Can the authors comment as to why none of the earlier purifications show this finding (Figure 1 for example)? Even more curious is that when YBAP1 is purified, YBX1 does not co-purify (Figure 4 supplement 1a, b).
14. Figure 4f, g - The text associated with these figures is very confusing, as is the labeling for the input. Also, what is "miR223 Fold change" in this regard? Seeing as your IgG should not have IP'd anything, normalizing to IgG can amplify noise. As such, RIP assays are typically presented as % input or fold enrichment.
15. Figure 4h - The authors show binding between miR223 and YBAP1 however it is not clear how significant this binding is. There is more than a 30-fold difference in binding affinity between miR223 and YBX1 than between miR223 and YBAP1. Even more, when comparing the EMSAs and fraction bound from figures 1 and 2 to those of Figure 4h, the binding between miR223 and YBAP1 more closely resembles that of miR190 and YBX1, which the authors state is a non-binder of YBX1. The authors will need to reconcile these discrepancies.
16. Can the authors present the Kd values for EMSA data?
17. Figure 5 - Does YBAP1-KO affect mitochondrial protein integrity or numbers?
18. Figure 6a - Are the authors using YBAP1 as their mitochondrial marker? Please include TOM20 and/or 22.
19. Figure 6b - Rab5 is an early endosome marker and may not fully represent the organelles that become MVBs. Co-localization at this point does not suggest that associating proteins will be present in the exosome, and it is possible that the authors are looking at the precursor of a recycling endosome. Even more, exosome loading does not occur at the early endosome, but instead at the MVB. Perhaps looking at markers of the late endosome such as Rab7 or ideally markers of the MVB such as M6P or CD63 would help draw an association between YBX1, YBAP1, and the exosome. Also, If the authors want to make the claim that interactions at the early endosome leads to secretion as an exosome, the authors should show that isolated EVs from Rab5Q79L-expressing cells contain miR223.
20. The mentioning of P-bodies is interesting but at no time is an association addressed. This is therefore an overly speculative conclusion. Either show an association or leave this out of the manuscript.
21. In lines 55-58, the authors make the comment "However, many of these studies used sedimentation at ~100,000 g to collect EVs, which may also collect RNP particles not enclosed within membranes which complicates the interpretation of these data." Do RNPs not dissolve when secreted? Can the authors give a reference for this statement? -

Reviewer #3 (Public Review):
The article by Ma et al pursues the previous work of the Schekman group, exploring the mechanisms of targeting of miRNAs into extracellular vesicles (EVs), or possibly exosomes, in HEK293 and U2OS cells. The authors had identified YBX1 as an RNA-binding protein required for the sorting of miR223 into CD63-expressing small EVs, probably mainly exosomes. Here they further observed that YBX1 directly binds miR223, which also binds to another protein, YBAP1, localized in mitochondria, where it sequesters miR223, thus preventing its targeting to MVBs' intraluminal vesicles. They observe the association of YBX1-containing P-bodies in the cytoplasm with mitochondria and with enlarged Rab5-endosomes and propose that this step is required for the exchange of miR223 for its loading into MVBs intraluminal vesicles and …
Reviewer #3 (Public Review):
The article by Ma et al pursues the previous work of the Schekman group, exploring the mechanisms of targeting of miRNAs into extracellular vesicles (EVs), or possibly exosomes, in HEK293 and U2OS cells. The authors had identified YBX1 as an RNA-binding protein required for the sorting of miR223 into CD63-expressing small EVs, probably mainly exosomes. Here they further observed that YBX1 directly binds miR223, which also binds to another protein, YBAP1, localized in mitochondria, where it sequesters miR223, thus preventing its targeting to MVBs' intraluminal vesicles. They observe the association of YBX1-containing P-bodies in the cytoplasm with mitochondria and with enlarged Rab5-endosomes and propose that this step is required for the exchange of miR223 for its loading into MVBs intraluminal vesicles and future exosomes.
The biochemical parts of the article, with quantitative experiments to decipher the molecular interactions of YBX1 and YBAP1 with miR223, are nicely performed and convincing. By contrast, the parts on the involvement of YBX1 and of YBAP1 in the release of miR223 in EVs or exosomes are more correlative than demonstrative and lack some controls. In particular, it is far-fetched to conclude from the observed movement (which may be serendipitous) of 2 P-bodies between mitochondria and enlarged endosomes (without any visualization of the miR) that this movement may be instrumental in the transfer of miR223 between mitochondria and putative exosomes (figures 6 and model in figure 7).
The experiments designed to evidence the mechanisms of miR223 release in EVs are also not sufficiently controlled and analysed to really support the interpretations. And the EV isolation steps are not performed in a way that supports the actual exosomal nature (i.e. exclusive origin from multivesicular endosome) of the EV analysed.
Another experimental weakness is that the authors make strong conclusions on MVBs and exosomes when they only analyse artificially-enlarged endosomes induced by overexpression of mutant Rab5. Although this approach has been used previously and shown CD63 in these induced enlarged compartments, it is an artificial blocking of normal endosomal trafficking, and may not reflect the situation of intracellular trafficking of miR223 in normal cells.
-