Single-cell analysis reveals dynamics of human B cell differentiation and identifies novel B and antibody-secreting cell intermediates
Curation statements for this article:-
Curated by eLife
eLife assessment
In this work, Verstegen and colleagues established an in vitro system and describe human B cell differentiation pathways via germinal center B cells towards plasma cells by performing single-cell analysis of in vitro stimulated human B cells. The study provides solid evidence toward establishment of in vitro model for B cell differentiation. This study may be valuable in differentiation of primary naive B cells into ASC ex vivo and will be of interest for immunologists with emphasis in B cell biology.
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Differentiation of B cells into antibody-secreting cells (ASCs) is a key process to generate protective humoral immunity. A detailed understanding of the cues controlling ASC differentiation is important to devise strategies to modulate antibody formation. Here, we dissected differentiation trajectories of human naive B cells into ASCs using single-cell RNA sequencing. By comparing transcriptomes of B cells at different stages of differentiation from an in vitro model with ex vivo B cells and ASCs, we uncovered a novel pre-ASC population present ex vivo in lymphoid tissues. For the first time, a germinal-center-like population is identified in vitro from human naive B cells and possibly progresses into a memory B cell population through an alternative route of differentiation, thus recapitulating in vivo human GC reactions. Our work allows further detailed characterization of human B cell differentiation into ASCs or memory B cells in both healthy and diseased conditions.
Article activity feed
-

-

Author Response
Reviewer #1 (Public Review):
[…] Overall, the results from these analyses are convincing and valuable, but still do not seem to be a big leap from their Unger 2021 paper […]. The methodology that they established should be described more clearly so that it can be shared with the research community. For example, they say cells how many donors were recruited for this experiment? are there differences in efficiency in B cell differentiation by individual?
Also, it would be important to assay for antibodies in the culture media. How would you suggest to improve the culture system to be used to model diseases?
We appreciate the reviewer's queries and the points raised. In response to the first set of comments, the reviewer has correctly observed that the methodology of the assay itself as employed in this paper is not …
Author Response
Reviewer #1 (Public Review):
[…] Overall, the results from these analyses are convincing and valuable, but still do not seem to be a big leap from their Unger 2021 paper […]. The methodology that they established should be described more clearly so that it can be shared with the research community. For example, they say cells how many donors were recruited for this experiment? are there differences in efficiency in B cell differentiation by individual?
Also, it would be important to assay for antibodies in the culture media. How would you suggest to improve the culture system to be used to model diseases?
We appreciate the reviewer's queries and the points raised. In response to the first set of comments, the reviewer has correctly observed that the methodology of the assay itself as employed in this paper is not new or superior to our previously published data in (Unger et al., Cells 2021), where we described a minimalistic in vitro system for efficient differentiation of human naive B cells into antibody-secreting cells (ASCs). However, the current study aims to elucidate a comprehensive evaluation of the phenotype of the cells in the in vitro system and their relationships in potential differentiation pathways. In addition, we aimed to elucidate how the detailed gene expression profiles of the differentiating cells in vitro compare to in vivo observed counterparts. In this way, we were able to uncover an antibody secreting cell phenotype in vivo that was not observed before and could only be uncovered due to our full transcriptome knowledge of these cells. In addition, we present novel findings that demonstrate that this culture system not only enables efficient ASCs generation but also recapitulates the entire in vivo B cell differentiation pathway, as evidenced by the presence of germinal-center (GC)-like and pre-memory B cells in the culture. These results have not been previously reported in the literature for human B cells in culture and represent a significant contribution to the field of human B cell biology.
In regards to the reviewer's inquiry about the cell culture protocol, its reproducibility, donors variability, and additional experimental applications, we refer to three additional recent publications from our group that have adopted the same in vitro B cell differentiation system and have provided extensive analysis of the immunoglobulin production, intracellular signaling pathways, as well as comparison with other culture systems in the field (Marsman et al., Cells 2020; Marsman et al., Eur. J. Immunol. 2022; Marsman et al., Front. Immunol. 2022). On top pf this, we now realize that the section that describes the culture system (MATERIAL AND METHODS - “In vitro naive B cell differentiation cultures”) was a bit too concise and we thank the reviewer for mentioning it. We have extended now on it and corrected an inconsistency at lines 125-127: “After six days, activated B cells were collected and co-cultured with 1 × 104 9:1 wild type (WT) to CD40L-expressing 3T3 cells that were irradiated and seeded one day in advance (as described above), together with IL-4 (100 ng/ml) and IL-21 (50 ng/ml; Invitrogen) for five days.”
As for the application of our in vitro system in disease modeling, as requested by the reviewer, this would require modifying the culture conditions to mimic the disease-specific biology background (if known). For instance, by inhibiting or enhancing specific transcriptional pathways that are known to be associated with the disease in question. However, it would also require the presence of antigen-specific B cells in the pool of naive B cells included in the culture, which can be difficult to achieve due to their low frequency. Alternatively, the system could be used to study antigen-specific recall responses using antigen-specific memory B cells as starting material. Our group has evaluated this approach in a recent publication (Marsman et al., Front. Immunol 2022).
[..] B cell differentiation may also influence to cell cycle regulation. Rather than normalize its effect, can authors analyze effect of cell cycle in B cell differentiation? [...]
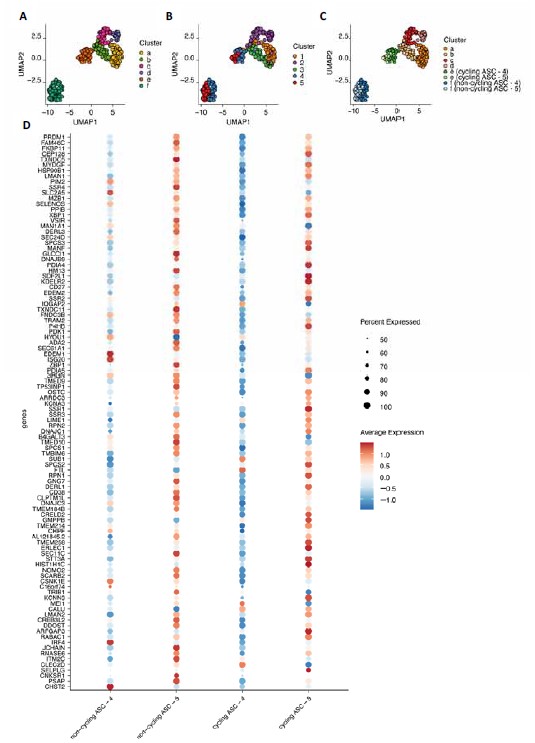
We very much agree with the reviewer and know that the cell cycle plays a significant role in B cell differentiation output trajectories (Zhou et al, Front Immunol. 2018; Duffy et al., Science 2012). Preparing the manuscript, we have in fact performed a parallel analysis in which we compared both cell cycle regressed- and not cell cycle regressed-based clustering and marker gene selection. Concerning the clustering, other clusters were obtained using the not cell-cycle-regressed dataset compared to the cell-cycle-regressed dataset (figure below). However, when overlaying the clusters obtained with the cell cycle-regressed dataset, the extra clusters were the same cell population but now split based on cycling and not cycling cells: cluster 2 is now divided into the cycling cluster “c”, and the not-cycling cluster “d” while cluster 4 and 5 are now divided into the cycling clusters “e” and the not-cycling cluster “f”. A comprehensive examination of the expression of the top 50 genes associated with antibody-secreting cells in the (non)cycling clusters 4 and 5 reveals that these genes are expressed at a higher level in (non)cycling cluster 5 as compared to cluster 4. This suggests that the cells within cluster 5 are more advanced in their differentiation, regardless of their cell cycle state. This finding has led us to the decision to present the data that has undergone cell cycle regression in the manuscript. Should the reviewer so desire, we are very willing to include additional supplementary figures to the manuscript that include the un-regressed representation.
Figure legend: A-C) UMAP projection of single-cell transcriptomes of in vitro differentiated human naive B cells without cell cycle regression. Each point represents one cell, and colors indicate graph-based cluster assignments identified without cell-cycle regression (A), with cell cycle regression (B) or with cell cycle regression and additional subdivision in cycling and not cycling cells (C). D) Dotplot showing the top 50 differentially expressed genes in cycling and not-cycling cells from cluster 4 and 5. Point size indicate percentage of cell in the cluster expressing the gene, color indicates average expression
-

eLife assessment
In this work, Verstegen and colleagues established an in vitro system and describe human B cell differentiation pathways via germinal center B cells towards plasma cells by performing single-cell analysis of in vitro stimulated human B cells. The study provides solid evidence toward establishment of in vitro model for B cell differentiation. This study may be valuable in differentiation of primary naive B cells into ASC ex vivo and will be of interest for immunologists with emphasis in B cell biology.
-

Reviewer #1 (Public Review):
The authors of this study used SMART-seq to study differentiating B cells. Then they performed extensive in silico analyses to validate that a subset of the cells mimicked human antibody-secreting cells. For example, they compared gene expression profile of each cluster in B cell developmental trajectory (Figs 1, 2), investigated gene enrichment in ASC-like cluster (Fig 3), adopted independent dataset (Fig 3), and compared gene expression signatures of their cells to those of GC ASCs (Fig 4). Overall, the results from these analyses are convincing and valuable, but still do not seem to be a big leap from their Unger 2021 paper and therefore making this study preliminary.
The methodology that they established should be described more clearly so that it can be shared with the research community. For example, …
Reviewer #1 (Public Review):
The authors of this study used SMART-seq to study differentiating B cells. Then they performed extensive in silico analyses to validate that a subset of the cells mimicked human antibody-secreting cells. For example, they compared gene expression profile of each cluster in B cell developmental trajectory (Figs 1, 2), investigated gene enrichment in ASC-like cluster (Fig 3), adopted independent dataset (Fig 3), and compared gene expression signatures of their cells to those of GC ASCs (Fig 4). Overall, the results from these analyses are convincing and valuable, but still do not seem to be a big leap from their Unger 2021 paper and therefore making this study preliminary.
The methodology that they established should be described more clearly so that it can be shared with the research community. For example, they say cells how many donors were recruited for this experiment? are there differences in efficiency in B cell differentiation by individual?
Also, it would be important to assay for antibodies in the culture media. How would you suggest to improve the culture system to be used to model diseases?
At the beginning the largest contributing factor for cell culstering was cell cycle. But B cell differentiation may also influence to cell cycle regulation. Rather than normalize its effect, can authors analyze effect of cell cycle in B cell differentiation? For example, identify sub-clusters shown in supple Fig 1g.
-

Reviewer #2 (Public Review):
In this work, Verstegen and colleagues try to delineate human B cell differentiation trajectories by using in vitro differentiation culture of human naive B cells. The authors adopted a protocol of B cell stimulation with CD40L-expressing fibroblasts and IL-4/IL-21, and cultured B cells were analyzed by single-cell transcriptome analysis. Five distinct clusters were identified with features of memory B cells, germinal center-like B cells, ASCs, pre-ASCs, or post-GC B cells. This work provides a precise description of gene expression profiles of activated B cell populations and some insight into the pathways of effector B cell differentiation. This work will be a solid basis for human B cell study using in vitro culture of target B cell populations, providing an excellent experimental protocol.
-