Quantitative proteomic analysis of skeletal muscles from wild-type and transgenic mice carrying recessive Ryr1 mutations linked to congenital myopathies
Curation statements for this article:-
Curated by eLife
eLife assessment
This paper provides a valuable systematic analysis of proteomic profiles associated with a particular murine Ryanodine receptor abnormality. Its analysis technique provides a solid and systematic set of data summarising the differences in different muscle types. The work emerges with insights into pathological mechanism of congenital muscle diseases linked to mutations in a range of other genes related to excitation contraction coupling in workers within the skeletal muscle field.
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Skeletal muscles are a highly structured tissue responsible for movement and metabolic regulation, which can be broadly subdivided into fast and slow twitch muscles with each type expressing common as well as specific sets of proteins. Congenital myopathies are a group of muscle diseases leading to a weak muscle phenotype caused by mutations in a number of genes including RYR1 . Patients carrying recessive RYR1 mutations usually present from birth and are generally more severely affected, showing preferential involvement of fast twitch muscles as well as extraocular and facial muscles. In order to gain more insight into the pathophysiology of recessive RYR1 -congential myopathies, we performed relative and absolute quantitative proteomic analysis of skeletal muscles from wild-type and transgenic mice carrying p.Q1970fsX16 and p.A4329D RyR1 mutations which were identified in a child with a severe congenital myopathy. Our in-depth proteomic analysis shows that recessive RYR1 mutations not only decrease the content of RyR1 protein in muscle, but change the expression of 1130, 753, and 967 proteins EDL, soleus and extraocular muscles, respectively. Specifically, recessive RYR1 mutations affect the expression level of proteins involved in calcium signaling, extracellular matrix, metabolism and ER protein quality control. This study also reveals the stoichiometry of major proteins involved in excitation contraction coupling and identifies novel potential pharmacological targets to treat RyR1-related congenital myopathies.
Article activity feed
-

-

Author Response
Reviewer #2 (Public Review):
- The main limitation of this study is that the results are primarily descriptive in nature, and thus, do not provide mechanistic insight into how Ryr1 disease mutations lead to the muscle-specific changes observed in the EDL, soleus and EOM proteomes.
An intrinsic feature of the high-throughput proteomic analysis technology is the generation of lists of differentially expressed proteins (DEP) in different muscles from WT and mutated mice. Although the definition of mechanistic insights related to changes of dozens of proteins is very interesting, it is a difficult task to accomplish and goes beyond the goal of the high-throughput proteomic analysis presented here. Nevertheless, the analysis of DEPs may indeed provide arguments to speculate on the pathogenesis of the phenotype linked to …
Author Response
Reviewer #2 (Public Review):
- The main limitation of this study is that the results are primarily descriptive in nature, and thus, do not provide mechanistic insight into how Ryr1 disease mutations lead to the muscle-specific changes observed in the EDL, soleus and EOM proteomes.
An intrinsic feature of the high-throughput proteomic analysis technology is the generation of lists of differentially expressed proteins (DEP) in different muscles from WT and mutated mice. Although the definition of mechanistic insights related to changes of dozens of proteins is very interesting, it is a difficult task to accomplish and goes beyond the goal of the high-throughput proteomic analysis presented here. Nevertheless, the analysis of DEPs may indeed provide arguments to speculate on the pathogenesis of the phenotype linked to recessive RyR1 mutations. In the unrevised manuscript, we pointed out that the fiber type I predominance observed in congenital myopathies linked to recessive Ryr1 mutation are consistent with the high expression level of heat shock proteins in slow twitch muscles. However, as suggested by Reviewer 3, we have removed "vague statements" from the text of the revised manuscript, concerning major insights into pathophysiological mechanisms, since we are aware that the mechanistic information, if any, that we can extract from the data set, cannot go over the intrinsic limitation of the high-throughput proteomic technology.
b) Results comparing fast twitch (EDL) and slow twitch (soleus) muscles from WT mice confirmed several known differences between the two muscle types. Similar analyses between EOM/EDL and EOM/soleus muscles from WT mice were not conducted.
We agree with the point raised by the Reviewer. In the revised manuscript we have changed Figure 2. The new Figure 2 shows the analysis of differentially expressed proteins in EDL, soleus and EOMs from WT mice. We have also added 2 new Tables (new Supplementary Table 2 and 3) and have inserted our findings in the revised Results section (page, 7, lines 157-176, pages 8 and 9).
c) While a reactome pathway analysis for proteins changes observed in EDL is shown in Supplemental Figure 1, the authors do not fully discuss the nature of the proteins and corresponding pathways impacted in the other two muscle groups analyzed.
We have now included in the revised manuscript a new Figure 2 which includes the Reactome pathway analysis comparing EDL with soleus, EDL with EOM and soleus with EOM (panels C, F and I, respectively). We have also inserted into the revised manuscript a brief description of the pathways showing the greatest changes in protein content (page 7 line 156-175, pages 8 and 9). We agree that the data showing changes in protein content between the 3 muscle groups of the WT mice are important also because they validate the results of the proteomic approach. Indeed, the present results confirm that many proteins including MyHCIIb, calsequestrin 1, SERCA1, parvalbumin etc are more abundantly expressed in fast twitch EDL muscles compared to soleus. Similarly, our results confirm that EOMs are enriched in MyHC-EO as well as cardiac isoforms of ECC proteins. This point has been clarified in the revised version of the manuscript (page 8, lines 198-213; page 9 lines 214-228). Nevertheless, we would like to point out that the main focus of our study is to compare the changes of protein content induced by the presence of recessive RyR1 mutations.
Reviewer #3 (Public Review):
a) it would be useful to determine whether changes in protein levels correlated with changes in mRNA levels …….
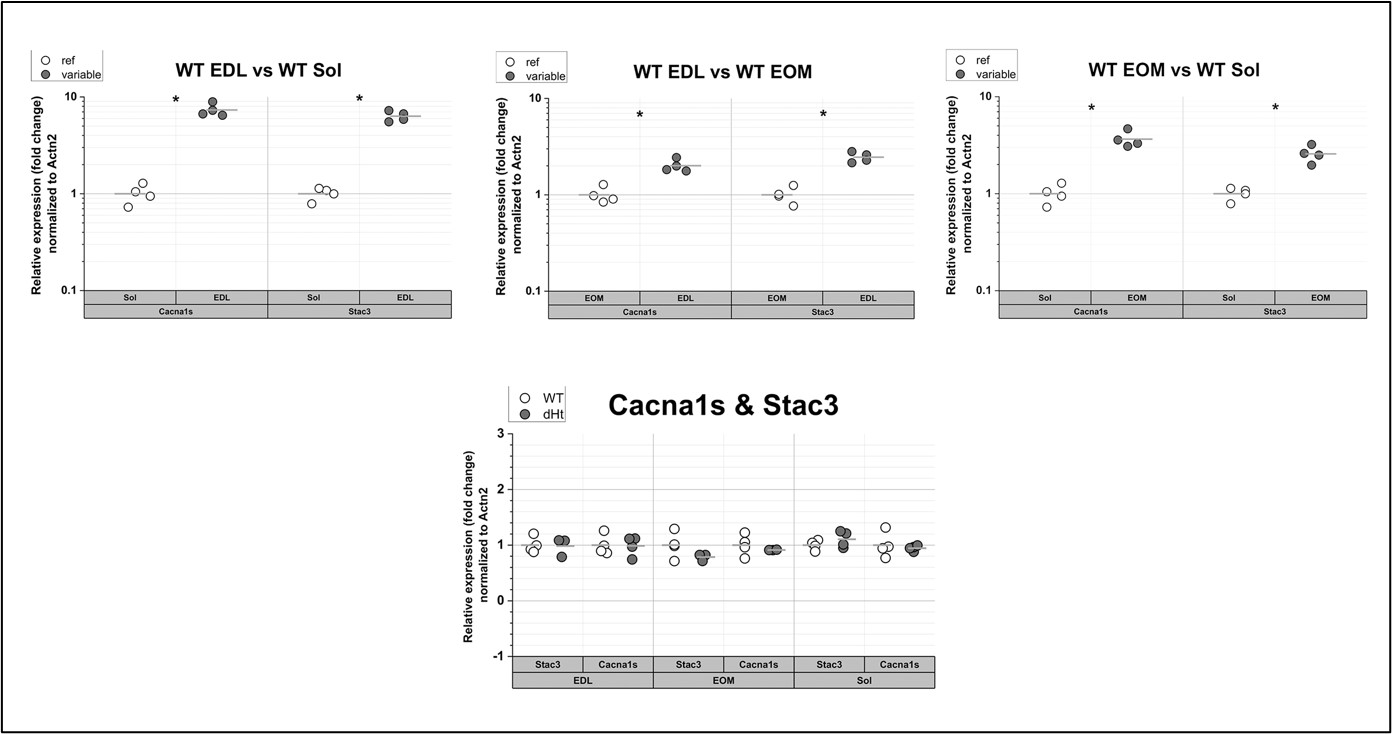
We performed qPCR analysis of Stac3 and Cacna1s in EDL, Soleus and EOM from WT mice (see Figure 1 below). The expression of transcripts encoding Cacna1s and Stac3 is approximately 9-fold higher in EDL compared to Soleus. The fold change of Stac3 and Cacna1s transcripts in EDL muscles is higher compared to the differences we observed by Mass spectrometry at the protein level between EDL and Soleus. Indeed, we found that the content of the Stac3 protein in EDL is 3-fold higher compared to that in soleus. Although there is no apparent linear correlation between mRNA and protein levels, we believe that a few plausible conclusions can be drawn, namely: (i) the expression level of both transcripts and proteins is higher EDL compared to EOM and soleus muscles, respectively, (ii) the expression level of transcripts encoding Stac3 correlate with those encoding Cacan1s and confirm proteomic data. In addition, the level of Stac3 transcript does not changes between WT and dHT, confirming our proteomic data which show that Stac3 protein content in muscles from dHT is similar to that found in WT littermates. Altogether these results support the concept that the differences in Stac3 content between EDL and soleus occur at both the protein and transcript levels, namely high Stac3 mRNA level correlates with higher protein content (EDL) and low mRNA levels correlated with low Stac3 protein content in Soleus muscles (see Figure 1 below).
Figure 2: qPCR of Cacna1s and Stac3 in muscles from WT mice. The expression levels of the transcripts encoding Cacna1s and Stac3 are the highest in EDL muscles and the lowest in soleus muscles (top panels). There are no significant changes in their relative expression levels in dHT vs WT. Each symbol represents the value from of a single mouse. * p=0.028 Mann Whitney test qPCR was performed as described in Elbaz et al., 2019 (Hum Mol Genet 28, 2987-2999).
….and whether or not the protein present was functional, and whether Stac3 was in fact stoichiometrically depleted in relation to Cacna1s.
We thought about this point but think that there are no plausible arguments to believe that Stac3 is not functional, one simple reason being that our WT mice do not have a phenotype which would be associated with the absence of Stac3 (Reinholt et al., PLoS One 8, e62760 2013, Nelson et al. Proc. Natl. Acad. Sci. USA 110:11881 2013).
b) In the abstract, the authors stated that skeletal muscle is responsible for voluntary movement. It is also responsible for non-voluntary. The abstract needs to be refocused on the mutation and on what we learn from this study. Please avoid vague statements like "we provide important insights to the pathophysiological mechanisms..." mainly when the study is descriptive and not mechanistic.
The abstract of the revised manuscript has been rewritten. In particular, we removed statements referring to important “pathophysiological mechanistic insight”.
c) The author should bring up the mutation name, location and phenotype early in the introduction.
In the revised manuscript we provide the information requested by the Reviewer (page 2 lines 36-38 and page 4, lines 98-102).
d) This reviewer also suggests that the authors refocus the introduction on the mutation location in the 3D RyR1 structure (available cryo-EM structure), if there is any nearby ligand binding site, protomers junction or any other known interacting protein partners. This will help the reader to understand how this mutation could be important for the channel's function
The residue Ala4329 is present inside the TMx (Auxiliary transmembrane helices) domain which spans from residue 4322 to 4370 and interposes structurally (des Georges A et al. 2016 Cell 167,145-57; Chen W, et al. 2020 EMBO Rep. 21, e49891). Although the structural resolution of the region has been improved (des Georges et al, 2016), parts of the domain still remain with no defined atomic coordinates, especially the region encompassing a.a. E4253 – F4540. Because of such undefined atomic coordinates of the region E4253-F4540, we are not able to determine the real orientation and the disposition of the amino acids in this region, including the A4329 residue. As reference, structure PDB: 5TAL of des Georges et al, 2016 was analyzed with UCSF Chimera (production version 1.16) (Pettersen et al. J. Comput. Chem. 25: 1605-1612. doi: 10.1002/jcc.20084).
-

eLife assessment
This paper provides a valuable systematic analysis of proteomic profiles associated with a particular murine Ryanodine receptor abnormality. Its analysis technique provides a solid and systematic set of data summarising the differences in different muscle types. The work emerges with insights into pathological mechanism of congenital muscle diseases linked to mutations in a range of other genes related to excitation contraction coupling in workers within the skeletal muscle field.
-

Reviewer #1 (Public Review):
This is a welcome contribution investigating proteomics in different physiological muscle types in a particular murine (DHT) Ryr1 abnormality. This recapitulates a particular human clinical condition. It emerges with a comparative analysis of the expression not only of RyR1 protein but also of other functional proteins. The work emerges with insights into pathological mechanism of congenital myopathies linked to mutations in a range of other genes related to excitation contraction coupling.
-

Reviewer #2 (Public Review):
This study used tandem mass isobaric tags (TMT) and LC-MS/MS analyses to complete proteomic analyses of whole extensor digitorum longus (EDL), soleus, and extraocular muscles (EOM) excised from 3 month old male WT (n=5) and dHT (n=5) mice. The major strengths of the work include the comprehensive nature of the unbiased muscle proteome studies, validation of the experimental approach by confirming several well-known differences between fast and slow twitch muscles in the WT EDL and soleus proteome data, and the identification of distinct proteome changes and alterations in core ECC and SOCE complex stoichiometry in the three different muscles from dHT mice. The main limitation of this study is that the results are primarily descriptive in nature, and thus, do not provide mechanistic insight into how Ryr1 …
Reviewer #2 (Public Review):
This study used tandem mass isobaric tags (TMT) and LC-MS/MS analyses to complete proteomic analyses of whole extensor digitorum longus (EDL), soleus, and extraocular muscles (EOM) excised from 3 month old male WT (n=5) and dHT (n=5) mice. The major strengths of the work include the comprehensive nature of the unbiased muscle proteome studies, validation of the experimental approach by confirming several well-known differences between fast and slow twitch muscles in the WT EDL and soleus proteome data, and the identification of distinct proteome changes and alterations in core ECC and SOCE complex stoichiometry in the three different muscles from dHT mice. The main limitation of this study is that the results are primarily descriptive in nature, and thus, do not provide mechanistic insight into how Ryr1 disease mutations lead to the muscle-specific changes observed in the EDL, soleus and EOM proteomes.
Results comparing fast twitch (EDL) and slow twitch (soleus) muscles from WT mice confirmed several known differences between the two muscle types (e.g. elevated type I myosin, slow troponin I/T/C isoforms, SERCA2, calsequestrin-2, and carbonic anhydrase 3 in soleus; elevated type IIb myosin, SERCA1, calsequestrin-1, collagen I, and parvalbumin in EDL), as well as an overall decrease in oxidoreductase activity associated proteins and increase in extracellular matrix proteins in EDL muscle. Relative levels of select proteins involved in muscle contraction, ECC, extracellular matrix, heat shock response, ribosomes, FK 506 binding, and calcium dependent kinase activity were are compared. Similar analyses between EOM/EDL and EOM/soleus muscles from WT mice were not conducted.
The authors next assessed changes in the EDL, soleus and EOM proteomes in muscles excised from dHT mice, which were previously shown to exhibit an early myopathy characterized by reductions in Ryr1 expression, muscle mass and specific force production. This analysis revealed that in addition to the expected decrease in Ryr1 levels in all three muscles, a large number of additional proteins were significantly increase/decreased altered in EDL (848 proteins), soleus (509 proteins), and EOM (677 proteins). Data in Fig. 3 indicate that more proteins were significantly upregulated than downregulated in all three dHT muscle groups. While a reactome pathway analysis for proteins changes observed in EDL is shown in Supplemental Figure 1, the authors do not fully discuss the nature of the proteins and corresponding pathways impacted in the other two muscle groups analyzed.
The authors conducted a targeted analysis of proteins involved in several select pathways known to be important for skeletal muscle (e.g. ECC proteins, contractile proteins, heat shock proteins, ribosomal proteins, FK506 binding proteins, calcium dependent protein kinases). Increases in some FK506 binding proteins were seen in EDL and EOM muscles of dHT mice, while increases in calcium dependent proteins kinases were observed in all three muscle groups. Overall, fewer protein changes were observed in soleus muscles of dHT mice, with most alterations impacting ECC and ribosomal proteins. Beyond the EDL reactome pathway analysis and author-selected protein analyses shown in Tables 2-4, the nature of the totality of proteins altered in each muscle group, the corresponding pathways involved, and the relative degree to which changes are conserved or unique across all three muscle groups analyzed are not fully evaluated or discussed.
The final part of this study used spiked-in labeled peptides in combination with parallel reaction-monitoring and high resolution TMT mass spectrometry to quantify several key proteins involved in coordinating the ECC (Ryr1 and Cacna1s) and SOCE (Stim1 and Orai1) processes. These analyses provide the first mass spectrometry-based quantification of the concentration (mol/kg) and stoichiometry (e.g. Ryr1/cacna1s, Stim1/Orai1, etc) of these proteins across the three different muscles in both WT and dHT mice. The results indicate that while the stoichiometry of the core ECC complex (Ryr1/Cacna1s ~0.6-0.7) is similar across all muscles in WT, this ratio is reduced in EDL and EOM (but not soleus) of dHT mice. Moreover, the stoichiometry of the core SOCE complex indicates that Orai1 levels are limiting in EDL and EOM muscle (Stim1/Orai1 ~25-50), while Orai1 protein was below detectable levels in soleus. Unlike the core ECC complex, core SOCE complex stoichiometry was unaltered in muscles of dHT mice. These findings have important implications regarding ECC and SOCE function in the three different muscle groups under both normal conditions and a mouse model of RYR1-related myopathy.
-

Reviewer #3 (Public Review):
The strength of this article is that the experiment performed was successfully validated by previously published results. However, it would be useful to determine whether changes in protein levels correlated with changes in mRNA levels and whether or not the protein present was functional, and whether Stac3 was in fact stoichiometrically depleted in relation to Cacna1s. The authors suggest that the change in RyR1 protein levels may have a knock-on effect on the levels of other proteins, which is a reasonable claim, but no experiments (such as using RNAi) were performed to confirm this. The authors also claim that an adaptive response exists to compensate for deleterious mutations, which is indeed well-established (see dosage compensation in x-linked disorders between XX women and XY men, for example), and …
Reviewer #3 (Public Review):
The strength of this article is that the experiment performed was successfully validated by previously published results. However, it would be useful to determine whether changes in protein levels correlated with changes in mRNA levels and whether or not the protein present was functional, and whether Stac3 was in fact stoichiometrically depleted in relation to Cacna1s. The authors suggest that the change in RyR1 protein levels may have a knock-on effect on the levels of other proteins, which is a reasonable claim, but no experiments (such as using RNAi) were performed to confirm this. The authors also claim that an adaptive response exists to compensate for deleterious mutations, which is indeed well-established (see dosage compensation in x-linked disorders between XX women and XY men, for example), and their experiment is consistent with this finding but does not itself show this on the level of cells, tissues, or the RyR itself.
Minor concerns.
- In the abstract, the authors stated that skeletal muscle is responsible for voluntary movement. It is also responsible for non-voluntary. The abstract needs to be refocused on the mutation and on what we learn from this study. Please avoid vague statements like "we provide important insights to the pathophysiological mechanisms..." mainly when the study is descriptive and not mechanistic.
- The author should bring up the mutation name, location and phenotype early in the introduction. This reviewer also suggests that the authors refocus the introduction on the mutation location in the 3D RyR1 structure (available cryo-EM structure), if there is any nearby ligand binding site, protomers junction or any other known interacting protein partners. This will help the reader to understand how this mutation could be important for the channel's function.
-