Molecular characterization of the intact mouse muscle spindle using a multi-omics approach
Curation statements for this article:-
Curated by eLife
eLife assessment
This manuscript is of potential interest for a broad spectrum of researchers working on the nervous and muscular systems. By combining transcriptome and proteome analyses, the authors reveal the molecular makeup of the different compartments of the muscle spindle. The work is novel, makes important observations, and is well-executed and methodologically convincing to provide the field with new tools for dissecting the development and function of the muscle spindle.
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
The proprioceptive system is essential for the control of coordinated movement, posture, and skeletal integrity. The sense of proprioception is produced in the brain using peripheral sensory input from receptors such as the muscle spindle, which detects changes in the length of skeletal muscles. Despite its importance, the molecular composition of the muscle spindle is largely unknown. In this study, we generated comprehensive transcriptomic and proteomic datasets of the entire muscle spindle isolated from the murine deep masseter muscle. We then associated differentially expressed genes with the various tissues composing the spindle using bioinformatic analysis. Immunostaining verified these predictions, thus establishing new markers for the different spindle tissues. Utilizing these markers, we identified the differentiation stages the spindle capsule cells undergo during development. Together, these findings provide comprehensive molecular characterization of the intact spindle as well as new tools to study its development and function in health and disease.
Article activity feed
-

-

Author Response
Reviewer #1 (Public Review):
Bornstein and colleagues address an important question regarding the molecular makeup of the different cellular compartments contributing to the muscle spindle. While work focusing on single components of the spindle in isolation - proprioceptors, gamma-motor neurons, and intrafusal muscle fibres - have been recently published, a comprehensive analysis of the transcriptome and proteome of the spindle was missing and it fills an important gap considering how local translation and protein synthesis can affect the development and function of such a specialised organ.
The authors combine bulk transcriptome and proteome analysis and identify new markers for neuronal, intrafusal, and capsule compartments that are validated in vivo and are shown to be useful for studying aspects of spindle …
Author Response
Reviewer #1 (Public Review):
Bornstein and colleagues address an important question regarding the molecular makeup of the different cellular compartments contributing to the muscle spindle. While work focusing on single components of the spindle in isolation - proprioceptors, gamma-motor neurons, and intrafusal muscle fibres - have been recently published, a comprehensive analysis of the transcriptome and proteome of the spindle was missing and it fills an important gap considering how local translation and protein synthesis can affect the development and function of such a specialised organ.
The authors combine bulk transcriptome and proteome analysis and identify new markers for neuronal, intrafusal, and capsule compartments that are validated in vivo and are shown to be useful for studying aspects of spindle differentiation during development. The methodology is sound and the conclusions in line with the results.
We thank the reviewer for highlighting the importance of our study.
I feel a bit more analysis regarding the specificity and developmental expression profiles of the identified markers would be a great addition. In particular:
- Are any of the proprioceptive sensory neurons markers specific for fibres innervating the muscle spindles or also found in Golgi tendon organs?
We thank the reviewer for the important question, following which we performed two additional analyses. First, in order to study the specificity of spindle afferent genes we identified, we examined the overlap between our list of 260 potential proprioceptive neuron genes and markers for the three proprioceptive neurons subtypes (Ia, II and Ib) identified by Wu and colleagues (Wu et al. 2021). As shown in the newly added Figure 1- figure supplement 2F, while we found many genes that are common to all subtypes, 69 genes exclusively overlapped with subtype markers (22 genes with type Ia neurons, 45 genes with type II neurons and 2 genes with both; lists are shown in Supplementary File 4). These results suggest that the 69 genes are expressed by muscle spindle afferents and not by GTO afferents.
Second, to study the specificity of our validated markers, we examined the expression of ATP1a3, VCAN and GLTU1, marking proprioception neurons, extracellular matrix and outer capsule, respectively, in GTOs. Results showed that all three markers were also detected in the different tissues composing the GTOs (newly added Figure 3 – figure supplement 3, below). As ATP1a3 is not in the 69 unique marker list, this analysis verified that it is expressed by all proprioceptive neurons. The expression of both VCAN and GLUT1 in GTO capsules highlights the similarity between the capsules of the two proprioceptors.
- On the same line are any of the gamma motor neurons markers found also in alpha?
We thank the reviewer for raising this issue. Following the reviewer’s question, we conducted a detailed analysis of the expression of potential γ motor neuron genes. To this end, we first generated a list of α-motor neurons genes in our data by performing ranked GSEA using published expression profiles of these neurons (Blum et al., 2021). Then, we compared between the three lists of neuronal genes, i.e. γ motor neurons, α motor neurons and proprioceptive neurons (newly added Figure 1 – figure supplement 2G), and found an overlap between the three lists. Nonetheless, we also identified 40 spindle genes that are specific to γ motor neuron (Figure 1 – figure supplement 2G and Supplementary File 4) and, therefore, are potential markers for these neurons.
- How early expression of ATP1A3 is found in neurons at the spindle or fibres starting to innervating the muscle? A couple of late embryonic timepoints would be great.
We thank the reviewer for this suggestion. We performed late embryonic (E15.5-E17.5) staining for ATP1a3, which showed its expression as early as E15.5 (new Figure 4 – figure supplement 1).
- Given that the approach used allows to obtain insights on whether local translation plays a major role into the differentiation of the spindle it would be interesting to assess whether the proprioceptor and gamma motor neuron markers identified are also found in the cell body or exclusively at the spindle.
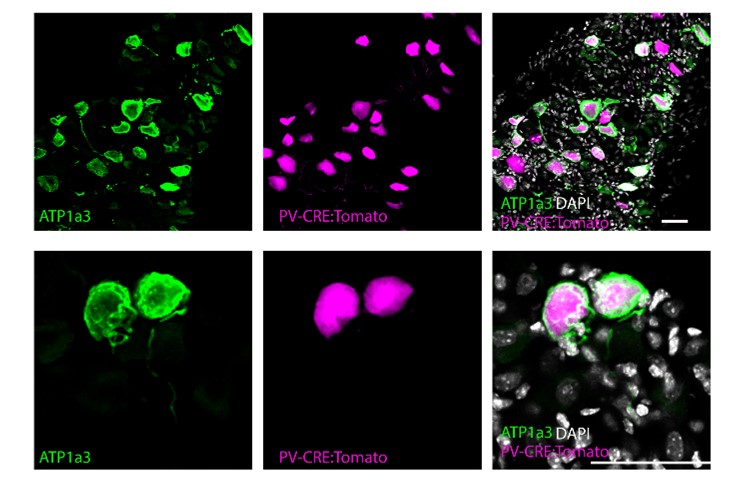
The reviewer raises an interesting question about local translation of the neuronal genes. Going through the literature, several lines of evidence indicate that the genes expressed at the neuronal end are also expressed in the neuron soma. In a study on retinal ganglion cell translatome, Holt and colleagues found that the axonal translatome is a subset of the significantly larger somal translatome (Shigeoka et al., Cell, 2016). Similarly, a study by Shuman and colleagues that compared the translatome of neuronal cell bodies, dendrites, and axons of rat hippocampal neurons showed that many common genes are translated, albeit at different levels (Glock et al., PNAS, 2021). Finally, following the reviewer’s suggestion, we studied the expression of ATP1a3 in the DRG, and found it to be expressed there as well (Figure L1). Thus, we predict that the markers we found in the neurons ends are likely also expressed in the soma. While this issue is very interesting, we believe that further validation of our assumption exceeds the scope of this study.
Figure L1. ATP1a3 expression in the DRG. Confocal images of DRG sections from adult PValb-Cre;tdTomato mice stained for ATP1a3 (magenta). Scale bars represent 50 μm.
Altogether, this is a novel and important work that will benefit scientists studying the neuromuscular and musculoskeletal systems by pushing the field toward an holistic understanding of the muscle spindle. These datasets in combination with the previous ones can be used to develop new genetic and viral strategies to study muscle spindle development and function in healthy and pathological states by analysing the roles and relative contributions of different components of this fascinating and still mysterious organ.
We thank again the reviewer for highlighting the importance of our study.
Reviewer #2 (Public Review):
The data presented are of high quality. Through complementary experiments involving the isolation of masseter muscle spindles, the authors perform RNA-seq and proteomic analysis, and identify genes and proteins that are differentially expressed in the muscle spindle versus the adjacent muscle fiber, and proteins that accumulate specifically in capsule cells and nerve endings. These data, while essentially descriptive, provide important information about the developmental framework of the sensory apparatus present in each muscle that accounts for its tension/contraction state. The data presented thus allow for a better characterization of muscle spindles and provide the community with a set of new markers for better identification of these structures. Analysis of the expression pattern of the Tomato reporter in transgenic animals under the control of Piezo2-CRE, Gli1-CRE and Thy1-YFP reporter reinforces the findings and the specificity of the expression pattern of the specific genes and proteins identified by the multi-omics approach and further validated by immunohistochemistry.
We thank the reviewer for the positive and encouraging feedback.
-

eLife assessment
This manuscript is of potential interest for a broad spectrum of researchers working on the nervous and muscular systems. By combining transcriptome and proteome analyses, the authors reveal the molecular makeup of the different compartments of the muscle spindle. The work is novel, makes important observations, and is well-executed and methodologically convincing to provide the field with new tools for dissecting the development and function of the muscle spindle.
-

Reviewer #1 (Public Review):
Bornstein and colleagues address an important question regarding the molecular makeup of the different cellular compartments contributing to the muscle spindle. While work focusing on single components of the spindle in isolation - proprioceptors, gamma-motor neurons, and intrafusal muscle fibres - have been recently published, a comprehensive analysis of the transcriptome and proteome of the spindle was missing and it fills an important gap considering how local translation and protein synthesis can affect the development and function of such a specialised organ.
The authors combine bulk transcriptome and proteome analysis and identify new markers for neuronal, intrafusal, and capsule compartments that are validated in vivo and are shown to be useful for studying aspects of spindle differentiation during …
Reviewer #1 (Public Review):
Bornstein and colleagues address an important question regarding the molecular makeup of the different cellular compartments contributing to the muscle spindle. While work focusing on single components of the spindle in isolation - proprioceptors, gamma-motor neurons, and intrafusal muscle fibres - have been recently published, a comprehensive analysis of the transcriptome and proteome of the spindle was missing and it fills an important gap considering how local translation and protein synthesis can affect the development and function of such a specialised organ.
The authors combine bulk transcriptome and proteome analysis and identify new markers for neuronal, intrafusal, and capsule compartments that are validated in vivo and are shown to be useful for studying aspects of spindle differentiation during development. The methodology is sound and the conclusions in line with the results. I feel a bit more analysis regarding the specificity and developmental expression profiles of the identified markers would be a great addition. In particular:
- Are any of the proprioceptive sensory neurons markers specific for fibres innervating the muscle spindles or also found in Golgi tendon organs?
- On the same line are any of the gamma motor neurons markers found also in alpha?
- How early expression of ATP1A3 is found in neurons at the spindle or fibres starting to innervating the muscle? A couple of late embryonic timepoints would be great.
- Given that the approach used allows to obtain insights on whether local translation plays a major role into the differentiation of the spindle it would be interesting to assess whether the proprioceptor and gamma motor neuron markers identified are also found in the cell body or exclusively at the spindle.
Altogether, this is a novel and important work that will benefit scientists studying the neuromuscular and musculoskeletal systems by pushing the field toward an holistic understanding of the muscle spindle. These datasets in combination with the previous ones can be used to develop new genetic and viral strategies to study muscle spindle development and function in healthy and pathological states by analysing the roles and relative contributions of different components of this fascinating and still mysterious organ.
-

Reviewer #2 (Public Review):
The data presented are of high quality. Through complementary experiments involving the isolation of masseter muscle spindles, the authors perform RNA-seq and proteomic analysis, and identify genes and proteins that are differentially expressed in the muscle spindle versus the adjacent muscle fiber, and proteins that accumulate specifically in capsule cells and nerve endings. These data, while essentially descriptive, provide important information about the developmental framework of the sensory apparatus present in each muscle that accounts for its tension/contraction state. The data presented thus allow for a better characterization of muscle spindles and provide the community with a set of new markers for better identification of these structures. Analysis of the expression pattern of the Tomato reporter …
Reviewer #2 (Public Review):
The data presented are of high quality. Through complementary experiments involving the isolation of masseter muscle spindles, the authors perform RNA-seq and proteomic analysis, and identify genes and proteins that are differentially expressed in the muscle spindle versus the adjacent muscle fiber, and proteins that accumulate specifically in capsule cells and nerve endings. These data, while essentially descriptive, provide important information about the developmental framework of the sensory apparatus present in each muscle that accounts for its tension/contraction state. The data presented thus allow for a better characterization of muscle spindles and provide the community with a set of new markers for better identification of these structures. Analysis of the expression pattern of the Tomato reporter in transgenic animals under the control of Piezo2-CRE, Gli1-CRE and Thy1-YFP reporter reinforces the findings and the specificity of the expression pattern of the specific genes and proteins identified by the multi-omics approach and further validated by immunohistochemistry.
-