Antiviral function and viral antagonism of the rapidly evolving dynein activating adaptor NINL
Curation statements for this article:-
Curated by eLife
Evaluation Summary:
This is an interesting discovery of a role for NINL in antiviral defense through modulation of interferon signaling. They found that there is diversifying selection of this factor as well as viral antagonism. This discovery paves the way to a better understanding of how viruses and hosts co-evolve.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #1 agreed to share their name with the authors.)
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Viruses interact with the intracellular transport machinery to promote viral replication. Such host–virus interactions can drive host gene adaptation, leaving signatures of pathogen-driven evolution in host genomes. Here, we leverage these genetic signatures to identify the dynein activating adaptor, ninein-like (NINL), as a critical component in the antiviral innate immune response and as a target of viral antagonism. Unique among genes encoding components of active dynein complexes, NINL has evolved under recurrent positive (diversifying) selection, particularly in its carboxy-terminal cargo-binding region. Consistent with a role for NINL in host immunity, we demonstrate that NINL knockout cells exhibit an impaired response to interferon, resulting in increased permissiveness to viral replication. Moreover, we show that proteases encoded by diverse picornaviruses and coronaviruses cleave and disrupt NINL function in a host- and virus-specific manner. Our work reveals the importance of NINL in the antiviral response and the utility of using signatures of host–virus genetic conflicts to uncover new components of antiviral immunity and targets of viral antagonism.
Article activity feed
-

-

Author Response
Reviewer #1 (Public Review):
This is a well performed study to demonstrate the antiviral function and viral antagonism of the dynein activating adapter NINL. The results are clearly presented to support the conclusions.
This reviewer has only one minor suggestion to improve the manuscript.
Add a discussion (1) why the folds of reduction among VSV, SinV and CVB3 were different in the NINL KO cells and (2) why the folds of reduction of VSV in the NINL KO A549 and U-2 OS cells.
Thank you for this suggestion. We have amended the results section to include additional information about these observations and possible explanations for these results.
Reviewer #2 (Public Review):
This manuscript is of interest to readers for host-viral co-evolution. This study has identified a novel human-virus interaction point …
Author Response
Reviewer #1 (Public Review):
This is a well performed study to demonstrate the antiviral function and viral antagonism of the dynein activating adapter NINL. The results are clearly presented to support the conclusions.
This reviewer has only one minor suggestion to improve the manuscript.
Add a discussion (1) why the folds of reduction among VSV, SinV and CVB3 were different in the NINL KO cells and (2) why the folds of reduction of VSV in the NINL KO A549 and U-2 OS cells.
Thank you for this suggestion. We have amended the results section to include additional information about these observations and possible explanations for these results.
Reviewer #2 (Public Review):
This manuscript is of interest to readers for host-viral co-evolution. This study has identified a novel human-virus interaction point NINL-viral 3C protease, where NINL is actively evolving upon the selection pressure against viral infect and viral 3Cpro cleavage. This study demonstrates that the viral 3Cpros-mediated cleavage of host NINL disrupts its adaptor function in dynein motor-mediated cargo transportation to the centrosome, and this disruption is both host- and virus-specific. In addition, this paper indicates the role of NINL in the IFN signaling pathway. Data shown in this manuscript support the major claims.
In this paper, the authors have identified a novel host-viral interaction, where viral 3C proteases (3Cpro) cleave at specific sites on a host activating adaptor of dynein intracellular transportation machinery, ninein-like protein (NINL or NLP in short) and inhibit its role in the antiviral innate immune response.
The authors firstly found that, unlike other activating adaptors of dynein intracellular transportation machinery, NINL (or NLP) is rapidly evolving. Thus, the authors hypothesized that this rapid evolution of NINL was caused by its interaction with viral infection. The authors found that viruses replicated higher in NINL knock-out (KO) cells than in wild-type (WT) cells and the replication level was not attenuated upon IFNa treatment in NINL KO cells, unlike in WT cells. Next, the authors investigated the role of NINL in type I IFN-mediated immune response and found that the induction of Janus kinase/signal transducer and activation of transcription (JAK/STAT) genes were attenuated in NINL KO cells upon IFNa treatment. The author further showed that the reduction of replication IFNa sensitive Vaccinia virus mutant upon IFNa treatment was decreased in NINL KO A549 cells compared to WT cells. The authors further showed that the virus antagonized NINL function by cleaving it with viral 3Cpro at its specific cleavage sites. NINL-peroxisome ligation-based cargo trafficking visualization assay showed that the redistribution of immobile membrane-bound peroxisome was disrupted by cleavage of NINL or viral infection.
This paper has revealed a novel host-virus interaction, and an antiviral function of a rapidly evolving activating adaptor of dynein intracellular transportation machinery, NINL. The major conclusions of this paper are well supported by data, but several aspects can be improved.
- It would be necessary to include a couple of other pathways involved in innate immune response besides JAK/STAT pathway.
We are very interested in this question as well. Our RNAseq data (Supplementary file 4 and Figure 3 – Figure supplement 4) suggest that there are several transcriptional changes that result from NINL KO. Our goal in this manuscript was to focus on IFN signaling in order to understand this specific effect of NINL KO since it might have wide-ranging consequences on viral replication. While we agree that broadening our studies to other signaling pathways, including other pathways involved in innate immune response, is a good idea, we feel that those experiments would take longer than two months to perform and therefore fall outside of the scope of this paper.
- The in-cell cleavages of NINL by viral 3Cpros were well demonstrated and supported by data of high quality. A direct biochemical demonstration of the cleavage is needed with purified proteins.
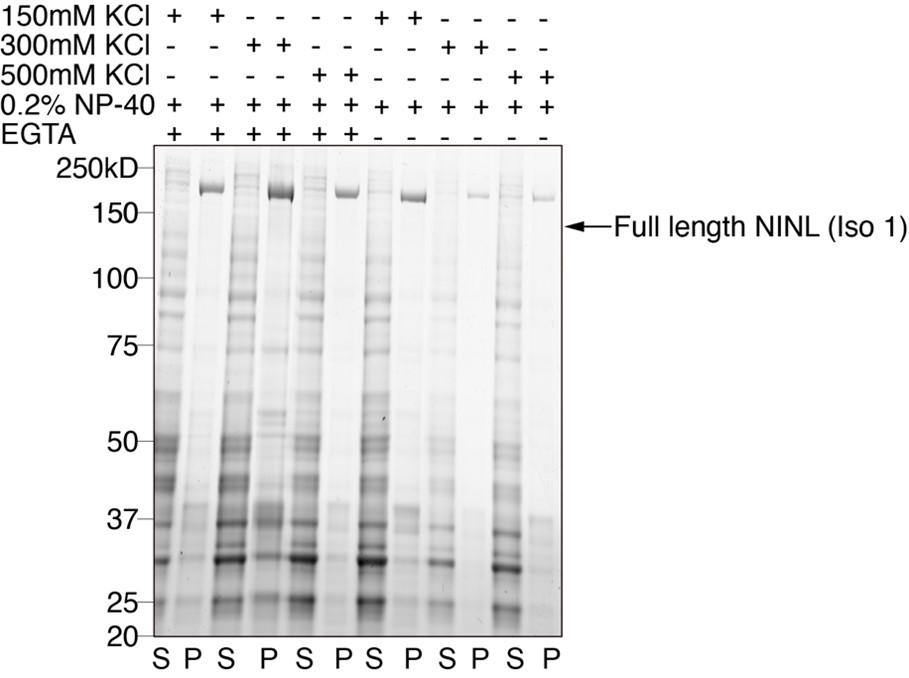
We agree with the reviewer that a direct biochemical cleavage assay would further demonstrate that viral 3Cpros cleave NINL specifically. However, our attempts to purify full-length NINL have been unsuccessful due to solubility issues (see example gel below), which is not surprising given that NINL is a >150 kDa human protein that has multiple surfaces that bind to other human proteins. As such, we focused our efforts on in-cell cleavage assays using specificity controls for cleavage. Specifically, we used catalytically inactive CVB3 3Cpro to show a dependence on protease catalytic activity and a variety of NINL constructs in which the glutamine in the P1 position is replaced by an arginine to show site specificity of cleavage. Notably, the cleavage sites in NINL that we mapped using this mutagenesis were predicted bioinformatically from known sites of 3Cpro cleavage in viral polyproteins, further indicating that cleavage is 3Cpro-dependent. We believe these results thus demonstrate that cleavage of NINL is dependent on viral protease activity and occurs in a sequence-specific manner. In light of the difficulty of purifying full-length NINL that would make biochemical experiments very challenging and likely take longer than two months to perform, we believe that our in cell data should be sufficient to demonstrate activity-dependent site-specific cleavage of NINL by viral 3Cpros.
Sypro stained SDS-PAGE gel showing supernatant (S) and insoluble pellet (P) fractions across multiple purifications with altered buffer conditions.
- The author used different cell types in different assays. Explain the rationale with a sentence for each assay.
Throughout this work, we choose to use a variety of cell lines for specific purposes. A549 cells were chosen as our main cell line as they are widely used in virology, are susceptible to the viruses we used, are responsive to interferon, and express both NINL and our control NIN at moderate levels. In the case of our virology and ISG expression data, we performed the same experiments with NINL KOs in other cell lines confirm that the phenotypes we observed in A549 cells could be attributed to the absence of NINL rather than off-target CRISPR perturbations or cell-line specific effects. All cleavage experiments were performed in HEK293T for their ease of transfection and protein expression. The inducible peroxisome trafficking assays were performed in U-2 OS cells as their morphology is ideal for observing the spatial organization of peroxisomes via confocal microscopy, and based on the fact that we had recapitulated the virology results and ISG expression results in those cells. At the suggestion of the reviewer, we have amended the text to include rationales where appropriate.
- While cell-based assays well support the conclusions in this paper, further demonstration in vivo would be helpful to provide an implication on the pathogenicity impact of NINL.
We agree. However, we believe that examining the impact of the loss of or antagonism of NINL on the pathogenesis of infectious diseases in an in vivo model is outside the scope of this study.
In summary, this manuscript contributes to a novel antiviral target. In addition, it is important to understand the host-virus co-evolution. The use of the evolution signatures to identify the "conflict point" between host and virus is novel.
-

Evaluation Summary:
This is an interesting discovery of a role for NINL in antiviral defense through modulation of interferon signaling. They found that there is diversifying selection of this factor as well as viral antagonism. This discovery paves the way to a better understanding of how viruses and hosts co-evolve.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #1 agreed to share their name with the authors.)
-

Reviewer #1 (Public Review):
This is a well performed study to demonstrate the antiviral function and viral antagonism of the dynein activating adapter NINL. The results are clearly presented to support the conclusions.
This reviewer has only one minor suggestion to improve the manuscript.
Add a discussion (1) why the folds of reduction among VSV, SinV and CVB3 were different in the NINL KO cells and (2) why the folds of reduction of VSV in the NINL KO A549 and U-2 OS cells.
-

Reviewer #2 (Public Review):
This manuscript is of interest to readers for host-viral co-evolution. This study has identified a novel human-virus interaction point NINL-viral 3C protease, where NINL is actively evolving upon the selection pressure against viral infect and viral 3Cpro cleavage. This study demonstrates that the viral 3Cpros-mediated cleavage of host NINL disrupts its adaptor function in dynein motor-mediated cargo transportation to the centrosome, and this disruption is both host- and virus-specific. In addition, this paper indicates the role of NINL in the IFN signaling pathway. Data shown in this manuscript support the major claims.
In this paper, the authors have identified a novel host-viral interaction, where viral 3C proteases (3Cpro) cleave at specific sites on a host activating adaptor of dynein intracellular …
Reviewer #2 (Public Review):
This manuscript is of interest to readers for host-viral co-evolution. This study has identified a novel human-virus interaction point NINL-viral 3C protease, where NINL is actively evolving upon the selection pressure against viral infect and viral 3Cpro cleavage. This study demonstrates that the viral 3Cpros-mediated cleavage of host NINL disrupts its adaptor function in dynein motor-mediated cargo transportation to the centrosome, and this disruption is both host- and virus-specific. In addition, this paper indicates the role of NINL in the IFN signaling pathway. Data shown in this manuscript support the major claims.
In this paper, the authors have identified a novel host-viral interaction, where viral 3C proteases (3Cpro) cleave at specific sites on a host activating adaptor of dynein intracellular transportation machinery, ninein-like protein (NINL or NLP in short) and inhibit its role in the antiviral innate immune response.
The authors firstly found that, unlike other activating adaptors of dynein intracellular transportation machinery, NINL (or NLP) is rapidly evolving. Thus, the authors hypothesized that this rapid evolution of NINL was caused by its interaction with viral infection. The authors found that viruses replicated higher in NINL knock-out (KO) cells than in wild-type (WT) cells and the replication level was not attenuated upon IFNa treatment in NINL KO cells, unlike in WT cells. Next, the authors investigated the role of NINL in type I IFN-mediated immune response and found that the induction of Janus kinase/signal transducer and activation of transcription (JAK/STAT) genes were attenuated in NINL KO cells upon IFNa treatment. The author further showed that the reduction of replication IFNa sensitive Vaccinia virus mutant upon IFNa treatment was decreased in NINL KO A549 cells compared to WT cells. The authors further showed that the virus antagonized NINL function by cleaving it with viral 3Cpro at its specific cleavage sites. NINL-peroxisome ligation-based cargo trafficking visualization assay showed that the redistribution of immobile membrane-bound peroxisome was disrupted by cleavage of NINL or viral infection.
This paper has revealed a novel host-virus interaction, and an antiviral function of a rapidly evolving activating adaptor of dynein intracellular transportation machinery, NINL. The major conclusions of this paper are well supported by data, but several aspects can be improved.
1. It would be necessary to include a couple of other pathways involved in innate immune response besides JAK/STAT pathway.
2. The in-cell cleavages of NINL by viral 3Cpros were well demonstrated and supported by data of high quality. A direct biochemical demonstration of the cleavage is needed with purified proteins.
3. The author used different cell types in different assays. Explain the rationale with a sentence for each assay.
4. While cell-based assays well support the conclusions in this paper, further demonstration in vivo would be helpful to provide an implication on the pathogenicity impact of NINL.In summary, this manuscript contributes to a novel antiviral target. In addition, it is important to understand the host-virus co-evolution. The use of the evolution signatures to identify the "conflict point" between host and virus is novel.
-