Macrophages regulate gastrointestinal motility through complement component 1q
Curation statements for this article:-
Curated by eLife
Evaluation Summary:
This study will be of broad interest to neuroscientists, immunologists, and gastroenterologists, revealing a novel role of complement C1q in intestinal macrophages and the regulation of gut motility. The manuscript is well written and key datasets are convincing, but the lack of mechanistic details that lead to dysmotility require additional supportive datasets at this time.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #3 agreed to share their name with the authors.)
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Peristaltic movement of the intestine propels food down the length of the gastrointestinal tract to promote nutrient absorption. Interactions between intestinal macrophages and the enteric nervous system regulate gastrointestinal motility, yet we have an incomplete understanding of the molecular mediators of this crosstalk. Here, we identify complement component 1q (C1q) as a macrophage product that regulates gut motility. Macrophages were the predominant source of C1q in the mouse intestine and most extraintestinal tissues. Although C1q mediates the complement-mediated killing of bacteria in the bloodstream, we found that C1q was not essential for the immune defense of the intestine. Instead, C1q-expressing macrophages were located in the intestinal submucosal and myenteric plexuses where they were closely associated with enteric neurons and expressed surface markers characteristic of nerve-adjacent macrophages in other tissues. Mice with a macrophage-specific deletion of C1qa showed changes in enteric neuronal gene expression, increased neurogenic activity of peristalsis, and accelerated intestinal transit. Our findings identify C1q as a key regulator of gastrointestinal motility and provide enhanced insight into the crosstalk between macrophages and the enteric nervous system.
Article activity feed
-

-

Author Response
Reviewer #3 (Public Review):
This manuscript by Pendse et al aimed to identify the role of the complement component C1q in intestinal homeostasis, expecting to find a role in mucosal immunity. Instead, however, they discovered an unexpected role for C1qa in regulating gut motility. First, using RNA-Seq and qPCR of cell populations isolated either by mechanical separation or flow cytometry, the authors found that the genes encoding the subunits of C1q are expressed predominantly in a sub-epithelial population of cells in the gut that Cd11b+MHCII+F4/80high, presumably macrophages. They support this conclusion by analyzing mice in which intestinal macrophages are depleted with anti-CSF1R antibody treatment and show substantial loss of C1qa, b and c transcripts. Then, they generate Lyz2Cre-C1qaflx/flx mice to genetically …
Author Response
Reviewer #3 (Public Review):
This manuscript by Pendse et al aimed to identify the role of the complement component C1q in intestinal homeostasis, expecting to find a role in mucosal immunity. Instead, however, they discovered an unexpected role for C1qa in regulating gut motility. First, using RNA-Seq and qPCR of cell populations isolated either by mechanical separation or flow cytometry, the authors found that the genes encoding the subunits of C1q are expressed predominantly in a sub-epithelial population of cells in the gut that Cd11b+MHCII+F4/80high, presumably macrophages. They support this conclusion by analyzing mice in which intestinal macrophages are depleted with anti-CSF1R antibody treatment and show substantial loss of C1qa, b and c transcripts. Then, they generate Lyz2Cre-C1qaflx/flx mice to genetically deplete C1qa in macrophages and assess the consequences on the fecal microbiome, transcript levels of cytokines, macromolecular permeability of the epithelial barrier, and immune cell populations, finding no major effects. Furthermore, provoking intestinal injury with chemical colitis or infection (Citrobacter) did not reveal macrophage C1qa-dependent changes in body weight or pathogen burden.
Then, they analyzed C1q expression by IHC of cross-sections of small and large intestine and find that C1q immunoreactivity is detectable adjacent to, but not colocalizing with, TUBB3+ nerve fibers and CD169+ cells in the submucosa. Interestingly, they find little C1q immunoreactivity in the muscularis externa. Nevertheless, they perform RNA-sequencing of LMMP preparations (longitudinal muscle with adherent myenteric plexus) and find a number of changes in gene ontology pathways associates with neuronal function. Finally, they perform GI motility testing on the conditional knockout mice and find that they have accelerated GI transit times manifesting with subtle changes in small intestinal transit and more profound changes in measures of colonic motility.
Overall, the manuscript is very well-written and the observation that macrophages are the major source of C1q in the intestine is well supported by the data, derived from multiple approaches. The observations on C1q localization in tissue and the strength of the conclusions that can be drawn from their conditional genetic model of C1qa depletion, however, would benefit from more rigorous validation.
- Interpretation of the majority of the findings in the paper rest on the specificity of the Lyz2 Cre for macrophages. While the specificity of this Cre to macrophages and some dendritic cells has been characterized in the literature in circulating immune cells, it is not clear if this has been characterized at the tissue level in the gut. Evidence demonstrating the selectivity of Cre activity in the gut would strengthen the conclusions that can be drawn.
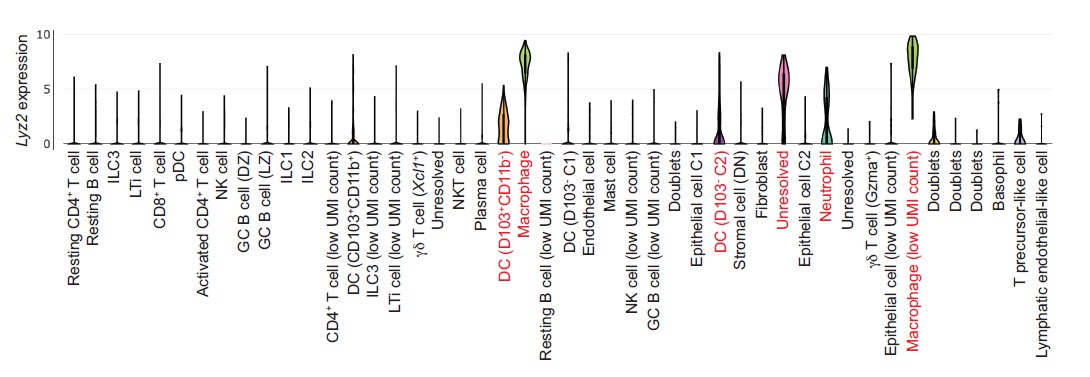
As indicated by the reviewer, Cre expression driven by the Lyz2 promoter is restricted to macrophages and some myeloid cells in the circulation (Clausen et al., 1999). To better understand intestinal Lyz2 expression at a cellular level, we analyzed Lyz2 transcripts from a published single cell RNAseq analysis of intestinal cells (Xu et al., 2019; see Figure below). These data show that intestinal Lyz2 is also predominantly expressed in gut macrophages with limited expression in dendritic cells and neutrophils.
Figure. Lyz2 expression from single cell RNAseq analysis of mouse intestinal cells. Data are from Xu et al., Immunity 51, 696-708 (2019). Analysis was done through the Single Cell Portal, a repository of scRNAseq data at the Broad Institute.
Additionally, our study shows that intestinal C1q expression is restricted to macrophages (CD11b+MHCII+F4/80hi) and is absent from other gut myeloid cell lineages (Figure 1E-H). This conclusion is supported by our finding that macrophage depletion via anti-CSF1R treatment also depletes most intestinal C1q (Figure 2A-C). Importantly, we found that the C1qaDMf mice retain C1q expression in the central nervous system (Figure 2 – figure supplement 1). Thus, the C1qaDMf mice allow us to assess the function of macrophage C1q in the gut and uncouple the functions of macrophage C1q from those of C1q in the central nervous system.
- Infectious and inflammatory colitis models were used to suggest that C1qa depletion in Lyz2+ lineage cells does not alter gut mucosal inflammation or immune response. However, the phenotyping of the mice in these models was somewhat cursory. For example, in DSS only body weight was shown without other typical and informative read-outs including colon length, histological changes, and disease activity scoring. Similarly, in Citrobacter only fecal cfu were measured. Especially if GI motility is accelerated in the KO mice, pathogen burden may not reflect efficiency of immune-mediated clearance alone.
We have added additional results which support our conclusion that C1qaDMf mice do not show a heightened sensitivity to acute chemically induced colitis. In Figure 3 – figure supplement 1 we now show a histological analysis of the small intestines of DSS-treated C1qafl/fl and C1qaΔMφ mice. This analysis shows that C1qaDMf mice have similar histopathology, colon lengths, and histopathology scores following DSS treatment. Likewise, our revised manuscript includes histological images of the colons of Citrobacter rodentium-infected C1qafl/fl and C1qaΔMφ mice showing similar pathology (Figure 3 – figure supplement 2).
- The evidence for C1q expression being restricted to nerve-associated macrophages in the submucosal plexus was insufficient. Localization was shown at low magnification on merged single-planar images taken from cross-sections. The data shown in Figure 4C is not of sufficient resolution to support the claims made - C1q immunoreactivity, for example, is very difficult to even see. Furthermore, nerve fibers closely approximate virtually type of macrophage in the gut, from those in the lamina propria to those in the muscularis….Finally, the resolution is too low to rule out C1q immunoreactivity in the muscularis externa.
Similar points were raised by Reviewer 2. Our original manuscript claimed that C1q-expressing macrophages were mostly located near enteric neurons in the submucosal plexus but were largely absent from the myenteric plexus. However, as both Reviewers have pointed out, this conclusion was based solely on our immunofluorescence analysis of tissue cross-sections.
To address this concern we further characterized C1q+ macrophage localization by performing a flow cytometry analysis on macrophages isolated from the mucosa (encompassing both the lamina propria and submucosa) and the muscularis, finding similar levels of C1q expression in macrophages from both tissues (Figure 4 – figure supplement 1 in the revised manuscript). Although the mucosal macrophage fraction encompasses both lamina propria and submucosal macrophages, our immunofluorescence analysis (Figure 4 B and C) suggests that the mucosal C1q-expressing macrophages are mostly from the submucosal plexus. This observation is consistent with the immunofluorescence studies of CD169+ macrophages shown in Asano et al., which suggest that most C169+ macrophages are located in or near the submucosal region, with fewer near the villus tips (Fig. 1e, Nat. Commun. 6, 7802).
Most importantly, our flow cytometry analysis indicates that the muscularis/myenteric plexus harbors C1q-expressing macrophages. To further characterize C1q expression in the muscularis, we performed RNAscope analysis by confocal microscopy of the myenteric plexus from mouse small intestine and colon (Figure 4D). The results show numerous C1q-expressing macrophages positioned close to myenteric plexus neurons, thus supporting the flow cytometry analysis. We note that although the majority of C1q immunofluorescence in our tissue cross-sections was observed in the submucosal plexus, we did observe some C1q expression in the muscularis by immunofluorescence (Figure 4B and C). We have rewritten the Results section to take these new findings into account.
Is the 5um average on the proximity analysis any different for other macrophage populations to support the idea of a special relationship between C1q-expressing macrophages and neurons?
We agree that the proximity analysis lacks context and have therefore removed it from the figure. The other data in the figure better support the idea that C1q+ macrophages are found predominantly in the submucosal and myenteric plexuses and that they are closely associated with neurons at these tissue sites.
There are many vessels in the submucosa and many associated perivascular nerve fibers - could the proximity simply reflect that both cell types are near vessels containing C1q in circulation?
Our revised manuscript includes RNAscope analysis showing C1q transcript expression by macrophages that are closely associated with enteric neurons (Figure 4D). These findings support the idea that the C1q close to enteric neurons is derived from macrophages rather than from the circulation.
- A major disconnect was between the observation that C1q expression is in the submucosa and the performance of RNA-seq studies on LMMP preparations. This makes it challenging to draw conclusions from the RNA-Seq data, and makes it particularly important to clarify the specificity of Lyz2-Cre activity.
Our revised manuscript provides flow cytometry data (Figure 4 – figure supplement 1) and RNAscope analysis (Figure 4D) showing that C1q is expressed in macrophages localized to the myenteric plexus. This accords with the results of our RNAseq analysis, which indicates altered LMMP neuronal function in C1qa∆Mφ mice (Figure 6A and B). Since neurons in the myenteric plexus are known to govern gut motility, it also helps to explain our finding that gut motility is accelerated in C1qa∆Mφ mice.
Finally, the pathways identified could reflect a loss of neurons or nerve fibers. No assessment of ENS health in terms of neuronal number or nerve fiber density is provided in either plexus.
Reviewers 1 and 2 also raised this point. Our revised manuscript includes a comparison of the numbers of enteric neurons in C1qafl/fl and C1qaΔMφ mice. There were no marked differences in neuron numbers in C1qaDMf mice when compared to C1qafl/fl controls (Figure 5A and B). There were also similar numbers of inhibitory (nitrergic) and excitatory (cholinergic) neuronal subsets and a similar enteric glial network (Figure 5C-E). Thus, our data suggest that the altered gut motility in the C1qaΔMφ mice arises from altered neuronal function rather than from an overt loss of neurons or nerve fibers. This conclusion is further supported by increased neurogenic activity of peristalsis (Figure 6H and I), and the expression of the C1q receptor BAI1 on enteric neurons (Figure 6 – figure supplement 4).
- To my knowledge, there is limited evidence that the submucosal plexus has an effect on GI motility. A recent publication suggests that even when mice lack 90% of their submucosal neurons, they are well-appearing without overt deficits (PMID: 29666241). Submucosal neurons, however, are well known to be involved in the secretomotor reflex and fluid flux across the epithelium. Assessment of these ENS functions in the knockout mice would be important and valuable.
Our revised manuscript provides new data showing C1q expression by muscularis macrophages in the myenteric plexus. We analyzed muscularis macrophages by flow cytometry and found that they express C1q (Figure 4 – figure supplement 1). These findings are further supported by RNAscope analysis of C1q expression in wholemounts of LMMP from small intestine and colon (Figure 4D and E). These results are thus consistent with the increased CMMC activity and accelerated gut motility in the C1qaDMf mice. As suggested by the reviewer, our finding of C1q+ macrophages in the submucosal plexus indicates that C1q may also have a role controlling the function of submucosal plexus neurons. We are further exploring this idea through extensive additional experimentation. Given the expanded scope of these studies, we are planning to include them in a follow-up manuscript.
- Immune function and GI motility can be highly sex-dependent - in all experiments mice of both sexes were reportedly used but it is not clear if sex effects were assessed.
This is a great point, and as suggested by the reviewer we indeed did encounter differences between male and female mice in our preliminary assays of gut motility. We therefore conducted our quantitative comparisons of gut motility between C1qafl/fl and C1qaDMf mice in male mice and now clearly indicate this point in the Materials and Methods.
-

Evaluation Summary:
This study will be of broad interest to neuroscientists, immunologists, and gastroenterologists, revealing a novel role of complement C1q in intestinal macrophages and the regulation of gut motility. The manuscript is well written and key datasets are convincing, but the lack of mechanistic details that lead to dysmotility require additional supportive datasets at this time.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #3 agreed to share their name with the authors.)
-

Reviewer #1 (Public Review):
The enteric nervous system (ENS) mediates gut motility and secretion, and recent work has shown that molecular crosstalk between enteric neurons and macrophages regulates ENS homeostasis and function. This study finds a novel role for complement C1q in this crosstalk. Complement plays a key role in immune defenses against pathogens, and has also been shown to play a key role in synaptic pruning in the central nervous system (CNS) during development, where it is expressed by microglial cells. However, the role of myeloid-derived complement components in ENS function is unknown. In this study, Hooper and colleagues reveal a novel role for C1q in mediating enteric neuron-macrophage crosstalk. First using targeted analysis of different cell-types, they reveal that C1q components (C1qa, C1qb, C1qc) are mainly …
Reviewer #1 (Public Review):
The enteric nervous system (ENS) mediates gut motility and secretion, and recent work has shown that molecular crosstalk between enteric neurons and macrophages regulates ENS homeostasis and function. This study finds a novel role for complement C1q in this crosstalk. Complement plays a key role in immune defenses against pathogens, and has also been shown to play a key role in synaptic pruning in the central nervous system (CNS) during development, where it is expressed by microglial cells. However, the role of myeloid-derived complement components in ENS function is unknown. In this study, Hooper and colleagues reveal a novel role for C1q in mediating enteric neuron-macrophage crosstalk. First using targeted analysis of different cell-types, they reveal that C1q components (C1qa, C1qb, C1qc) are mainly expressed by subepithelial immune cells, and in particular intestinal macrophages (CD11b+MHCII-F4/80hi). Elimination of macrophages using anti-CSFR1 led to decreased C1q. C1qafl/fl conditional mice were crossed with LysM-Cre to eliminate its expression in myeloid immune cells. This led to loss of C1q in the gut as well as other peripheral tissues but not the brain. Next, the authors find that loss of C1qa from macrophages led to no changes in microbiome composition, gut barrier permeability, susceptibility to DSS induced colitis, or Citrobacter rodentium infection. The immune populations in the gut were also not majorly affected. Imaging showed that C1q macrophages were mainly in submucosal plexus and located next to nerves. RNAseq showed changes in neuronal gene pathways related to synaptic organization in the associated tissues from C1q macrophage KO mice. Functionally, the authors then found that several measures of gut motility showed dysregulation, with faster transit time by carmine red, rhodamine B transit assay, a peristalsis assay, and colonic bead expulsion. Thus macrophage expressed C1q regulates peristalsis and neuronal function in the gut.
This is an exciting study that reveals a novel role for complement in regulating macrophage-neuron crosstalk and gut motility. Strengths include the use of interdisciplinary assays to first identify the cell-type that produces C1q, and then determine that this loss does not affect certain measures (microbiome composition, colitis, infection susceptibility), but does affect other measures (neuronal gene expression and gut motility measures). One remaining question is how C1q affects the nervous system, and a better characterization of neuronal composition or structure would be needed to rule in or rule out a mechanism.
-

Reviewer #2 (Public Review):
1. The manuscript is well-written, the data is of high quality and presented in a very logical manner.
2. The data demonstrating the motility defect in Lyz2-Cre:C1q-fl/fl mice is convincing.
3. The results supporting the conclusion that intestinal macrophages are the major source of C1q are strong.
4. The conclusion that C1q is mostly expressed by ENS-associated muscularis macrophages but not mucosal macrophages is not fully supported by the data.
5. Although comparison of the gene expression in Lyz2-Cre:C1q-fl/fl mice and control mice predicts changes in the ENS development and/or function, there is no data that would identify an ENS cell type(s) directly targeted by C1q and demonstrate their functional change (either in vivo or in vitro). This makes the conclusion of the direct link between …Reviewer #2 (Public Review):
1. The manuscript is well-written, the data is of high quality and presented in a very logical manner.
2. The data demonstrating the motility defect in Lyz2-Cre:C1q-fl/fl mice is convincing.
3. The results supporting the conclusion that intestinal macrophages are the major source of C1q are strong.
4. The conclusion that C1q is mostly expressed by ENS-associated muscularis macrophages but not mucosal macrophages is not fully supported by the data.
5. Although comparison of the gene expression in Lyz2-Cre:C1q-fl/fl mice and control mice predicts changes in the ENS development and/or function, there is no data that would identify an ENS cell type(s) directly targeted by C1q and demonstrate their functional change (either in vivo or in vitro). This makes the conclusion of the direct link between macrophage-derived C1q and regulation of the ENS function questionable. -

Reviewer #3 (Public Review):
This manuscript by Pendse et al aimed to identify the role of the complement component C1q in intestinal homeostasis, expecting to find a role in mucosal immunity. Instead, however, they discovered an unexpected role for C1qa in regulating gut motility. First, using RNA-Seq and qPCR of cell populations isolated either by mechanical separation or flow cytometry, the authors found that the genes encoding the subunits of C1q are expressed predominantly in a sub-epithelial population of cells in the gut that Cd11b+MHCII+F4/80high, presumably macrophages. They support this conclusion by analyzing mice in which intestinal macrophages are depleted with anti-CSF1R antibody treatment and show substantial loss of C1qa, b and c transcripts. Then, they generate Lyz2Cre-C1qaflx/flx mice to genetically deplete C1qa in …
Reviewer #3 (Public Review):
This manuscript by Pendse et al aimed to identify the role of the complement component C1q in intestinal homeostasis, expecting to find a role in mucosal immunity. Instead, however, they discovered an unexpected role for C1qa in regulating gut motility. First, using RNA-Seq and qPCR of cell populations isolated either by mechanical separation or flow cytometry, the authors found that the genes encoding the subunits of C1q are expressed predominantly in a sub-epithelial population of cells in the gut that Cd11b+MHCII+F4/80high, presumably macrophages. They support this conclusion by analyzing mice in which intestinal macrophages are depleted with anti-CSF1R antibody treatment and show substantial loss of C1qa, b and c transcripts. Then, they generate Lyz2Cre-C1qaflx/flx mice to genetically deplete C1qa in macrophages and assess the consequences on the fecal microbiome, transcript levels of cytokines, macromolecular permeability of the epithelial barrier, and immune cell populations, finding no major effects. Furthermore, provoking intestinal injury with chemical colitis or infection (Citrobacter) did not reveal macrophage C1qa-dependent changes in body weight or pathogen burden.
Then, they analyzed C1q expression by IHC of cross-sections of small and large intestine and find that C1q immunoreactivity is detectable adjacent to, but not colocalizing with, TUBB3+ nerve fibers and CD169+ cells in the submucosa. Interestingly, they find little C1q immunoreactivity in the muscularis externa. Nevertheless, they perform RNA-sequencing of LMMP preparations (longitudinal muscle with adherent myenteric plexus) and find a number of changes in gene ontology pathways associates with neuronal function. Finally, they perform GI motility testing on the conditional knockout mice and find that they have accelerated GI transit times manifesting with subtle changes in small intestinal transit and more profound changes in measures of colonic motility.
Overall, the manuscript is very well-written and the observation that macrophages are the major source of C1q in the intestine is well supported by the data, derived from multiple approaches. The observations on C1q localization in tissue and the strength of the conclusions that can be drawn from their conditional genetic model of C1qa depletion, however, would benefit from more rigorous validation.
1 - Interpretation of the majority of the findings in the paper rest on the specificity of the Lyz2 Cre for macrophages. While the specificity of this Cre to macrophages and some dendritic cells has been characterized in the literature in circulating immune cells, it is not clear if this has been characterized at the tissue level in the gut. Evidence demonstrating the selectivity of Cre activity in the gut would strengthen the conclusions that can be drawn.
2 - Infectious and inflammatory colitis models were used to suggest that C1qa depletion in Lyz2+ lineage cells does not alter gut mucosal inflammation or immune response. However, the phenotyping of the mice in these models was somewhat cursory. For example, in DSS only body weight was shown without other typical and informative read-outs including colon length, histological changes, and disease activity scoring. Similarly, in Citrobacter only fecal cfu were measured. Especially if GI motility is accelerated in the KO mice, pathogen burden may not reflect efficiency of immune-mediated clearance alone.
3 - The evidence for C1q expression being restricted to nerve-associated macrophages in the submucosal plexus was insufficient. Localization was shown at low magnification on merged single-planar images taken from cross-sections. The data shown in Figure 4C is not of sufficient resolution to support the claims made - C1q immunoreactivity, for example, is very difficult to even see. Furthermore, nerve fibers closely approximate virtually type of macrophage in the gut, from those in the lamina propria to those in the muscularis. Is the 5um average on the proximity analysis any different for other macrophage populations to support the idea of a special relationship between C1q-expressing macrophages and neurons? There are many vessels in the submucosa and many associated perivascular nerve fibers - could the proximity simply reflect that both cell types are near vessels containing C1q in circulation? Finally, the resolution is too low to rule out C1q immunoreactivity in the muscularis externa.
4 - A major disconnect was between the observation that C1q expression is in the submucosa and the performance of RNA-seq studies on LMMP preparations. This makes it challenging to draw conclusions from the RNA-Seq data, and makes it particularly important to clarify the specificity of Lyz2-Cre activity. Finally, the pathways identified could reflect a loss of neurons or nerve fibers. No assessment of ENS health in terms of neuronal number or nerve fiber density is provided in either plexus.
5 - To my knowledge, there is limited evidence that the submucosal plexus has an effect on GI motility. A recent publication suggests that even when mice lack 90% of their submucosal neurons, they are well-appearing without overt deficits (PMID: 29666241). Submucosal neurons, however, are well known to be involved in the secretomotor reflex and fluid flux across the epithelium. Assessment of these ENS functions in the knockout mice would be important and valuable.
5 - Immune function and GI motility can be highly sex-dependent - in all experiments mice of both sexes were reportedly used but it is not clear if sex effects were assessed.
-