Structure of Dunaliella photosystem II reveals conformational flexibility of stacked and unstacked supercomplexes
Curation statements for this article:-
Curated by eLife
eLife assessment
The work of Caspy and coworkers resolves the cryo-EM structures of stacked and unstacked PSII supercomplexes of Dunaliella, revealing unexpected connectivity and conformational flexibility, with intriguing implications for the function and regulation of photosynthesis.
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Photosystem II (PSII) generates an oxidant whose redox potential is high enough to enable water oxidation , a substrate so abundant that it assures a practically unlimited electron source for life on earth . Our knowledge on the mechanism of water photooxidation was greatly advanced by high-resolution structures of prokaryotic PSII . Here, we show high-resolution cryogenic electron microscopy (cryo-EM) structures of eukaryotic PSII from the green alga Dunaliella salina at two distinct conformations. The conformers are also present in stacked PSII, exhibiting flexibility that may be relevant to the grana formation in chloroplasts of the green lineage. CP29, one of PSII associated light-harvesting antennae, plays a major role in distinguishing the two conformations of the supercomplex. We also show that the stacked PSII dimer, a form suggested to support the organisation of thylakoid membranes , can appear in many different orientations providing a flexible stacking mechanism for the arrangement of grana stacks in thylakoids. Our findings provide a structural basis for the heterogenous nature of the eukaryotic PSII on multiple levels.
Article activity feed
-

-

Author Response
Reviewer #2 (Public Review):
I am not a specialist in cryo-EM, so cannot comment on the technicalities of the structure reconstruction or methods used. I thus focus on the conclusions and observations that the authors provide in the manuscript and their relevance to functional photosynthesis.
The authors attempt to resolve the structure of PSII from Dunaliella and noticed that three types of PSII could be identified: two conformational states, and a stacked configuration. There is no doubt that these structures add to our current knowledge of PSII and that they exist in abundance upon solubilisation of the sample. My main issue however is the relevance to in vivo conditions, and the efforts to exclude the possibility that pigment loss and conformational states and stacking are a reflection of ex-vivo manipulations.
Author Response
Reviewer #2 (Public Review):
I am not a specialist in cryo-EM, so cannot comment on the technicalities of the structure reconstruction or methods used. I thus focus on the conclusions and observations that the authors provide in the manuscript and their relevance to functional photosynthesis.
The authors attempt to resolve the structure of PSII from Dunaliella and noticed that three types of PSII could be identified: two conformational states, and a stacked configuration. There is no doubt that these structures add to our current knowledge of PSII and that they exist in abundance upon solubilisation of the sample. My main issue however is the relevance to in vivo conditions, and the efforts to exclude the possibility that pigment loss and conformational states and stacking are a reflection of ex-vivo manipulations.
Our compact model contains 202 Chls molecules while the stretched conformation contains 206 Chls. All of the differences in Chl binding are attributed to CP29. We have compiled a table enumerating the different CP29 structures currently available from plants and green alga at similar resolution to our work (Supplementary table 2). In the larger plant complexes (C2S2M2) CP29 contains 14 chls, while CP29 in smaller C2S2 complexes contains 10-13 chls, so it appears the some chl loss from CP29 is associated with the release of LHCIIM. In the green alga structures, CP29 contains less chls in general and shows a similar trend. The currently published structure most relevant to our work contains 8 chls (6KAC), a somewhat lower amount then both the compact and stretched models (9 and 11 chls, respectively). The stretched orientation, which is the closest match to the known PSII core arrangement, therefore contains more chls than comparable models. While the in-vivo configuration is not known in the sense that it could contain more chls, the current structure is apparently the closest representation of it.
The presence of CP29 with lower chls content in the chlamy C2S2 (6KAC, which is in a stretched orientation) supports a conclusion that pigment loss from CP29 alone is not sufficient to trigger the stretch to compact transition although it is associated with it. In general, the precise orientation of CP29 is variable and seem to depend on the binding of additional LHCII, it is possible that some chl loss is accompanied with these changes in vivo.
I see a number of questions pertaining to this work. Starting from the two conformations of PSII, compact and stretched, the authors say that both are highly active based on oxygen measurements at a saturating light intensity. In the meantime, they report large variations in the chl content and positions of the chlorophyll molecules in these structures (also compared to other known PSIIs). This gives the impression that one can lose two chlorophylls, and freely modify the distance between others without losing efficiency, certainly a risky conclusion. Are the samples highly active also in light-limiting conditions? It is thought that even tiny movements and alterations in chl-chl distances alter their coupling and spectral properties, how come the variations in this report are so huge? In other words, the assay tests the charge separation activity of the PSII RC in the preps, but not the light-harvesting efficiency.
The chl content differences reported in this work amounts to 2%. In our opinion this represents quite a low variation in pigment content, which exist in virtually any experiment involving large complexes. We agree that measurements of activity in limiting light conditions are interesting, however this goes beyond the scope of the current work. Light harvesting efficiency in PSII is known to vary substantially as a result of additional mechanisms (NPQ in some of its forms), not associated with chl loss or gain. While the formation of quenching centers is attributed to small structural changes within specific pigment protein complexes, what we are showing in this work are structural changes between pigment protein complexes. These can affect transfer rates between the different complexes but are distinct from the structural changes thought to accompany the formation of quenching centers within specific pigment protein complexes.
How does one ascertain that the lost chlorophyll molecules in CP29 are not a preparation error? Does slightly increasing the detergent concentration impact the proportion of stretched:compact forms?
The effect of detergent concentration on the proportion of the different forms was not tested directly. However, we do not detect many differences in lipids or bound detergent molecules content between the two conformations, suggesting that for these “ligands” the differences are not substantial. We can only distinguish these two forms at the very last stages of data processing, at the present state of cryoEM cost and time availability, mapping the effect of detergent concentration on the different orientations is outside our reach.
On a similar note, how do the authors exclude that a certain interaction with this type of grid impacts the distribution of these complexes? Is it identical to a biologically separate preparation of algae? In case of discoveries of this type, it is of high importance to exclude as many possibilities of non-native conditions or influences on the structure.
It’s hard to completely exclude grid and sample preparation issues. However, we employed relatively standard grids and vitrification conditions. The observed complexes are embedded in vitrified ice and do not interact with the grid directly. The differences we observed are mainly in the orientations of the PSII cores, all the interactions between PSII subunits within each core are preserved and agree with previously published structures. Since the interactions within the core and between cores involve the same physical principles, we think its fairly conservative to think that the observed core orientations are not an artefact of sample preparation.
I would further like to encourage the authors to elaborate on the CP29 phosphorylation. What is the proportion of PSIIcomp that are phosphorylated? I assume it is not 100%, as in this case, the authors would propose that this is the effect that modulates between compact and stretched architectures.
Its difficult to estimate the proportion of observed phosphorylation/sulfinylation. To be detected in maps, most of the residues (above 50%) are probably modified. We attempted to estimate this by refining the atom occupancies of the Pi molecule on Ser84 and the oxygens attached to Cys218, both values suggested that about 70% of the complexes are modified. With regards to the possibility that these modifications can promote the formation of the compact state, we think that this is certainly a possibility, since these modifications were detected in this state and are in close proximity to each other. However, this can also result from the resolution differences of the maps and the structural implications of both modifications are hard to predict. At this point we prefer to note their existence without further interpretations.
In line 290, the authors highlight the structural heterogeneity within the two groups' PSII conformations. I would like to see how does the distribution look like for all the structures together: are the two (stretched and compact) specifically forming two heterogenous distributions? Or is it possible that the distribution between the two is quasi-continuous? In other words, if the structures are not perfectly defined, how do the authors decide that two- and not more or less subtypes exist?
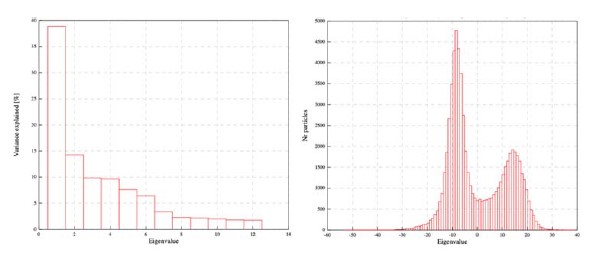
We went back and refined the initial particle group (containing both compact and stretched orientations) using multibody with masks defining the two PSII monomers. This analysis showed the expected two peaks only in the first Principal components which accounted for ~38% of the variance in the dataset.
Multibody refinement carried out on the combined particle dataset shows one very large PC accounting for about 38% of the variance and the presence of two distinct peaks in the particle distribution of the first PC.
From this analysis it’s clear that there are two distinct classes in this particle set (as expected), as none of the other PC’s shows any signs of multiple peaks, this analysis suggests that two distinct models are the best representation of this eukaryotic PSII. Whether these are quasi continuous or distinct is more complex. There is continuity in this representation (particle distributions along PC), a different picture may appear if characters such as CP29 state are considered, but the size of CP29 and the remaining heterogeneity does not provide enough signal to carry out this classification at the moment.
Considering the stacked PSII, I also have a few concerns. Contrary to previous studies the authors do not assign a functional role to the stacking beyond the structural aspect. This could be better backed by a discussion about the closest chlorophyll a molecules across the stacked PSII, which given the rather large distance shown in fig. 4L seems to be too large for any EET across the stromal gap.
The closest chl-chl distance that we can measure in the stacked PSII dimer is ~54 Å, with most distances at the ~70 Å range, making EET between staked complexes very slow. We have added a statement clarifying this to our manuscript. In our opinion a structural role for the staked PSII dimer is more likely.
There is a report that suggests the presence of some density between the stacked PSII - could the authors comment on the differences between it and their work? Are the angles and positions conserved between these types of stacks? https://doi.org/10.1038/s41598-017-10700-8
We referred to Albanese et al, in our manuscript. We isolated the C2S2 complex from green alga, the analysis in Albanese et al was done on C2S2M1 complexes from pea and this can account for some of the differences. At any rate, our conclusion that we don’t find any evidence for protein linkers in the stacked complex is stated clearly. The angles described in Albanese et al are consistent with our analysis.
Line 387, the authors state that due to the transient nature of the interactions across the stromal gap, the stacks could be "under-detected" in cryo-ET data. This statement is in my opinion misformulated. For once, the transient interaction argument would apply the same (if not more due to changing conditions induced by the purification process) to the single particle analysis performed in this paper. Second, tomographic volumes detect hundreds of PSII in a suspended state. Any transient interaction that adds up to 25% of particle population in a steady state cell should be clearly visible, while the in situ data suggests not more than random cross-stromal-gap orientations. Of course, this can be a specificity of Chlamydomonas or a particular growth condition. The statement used by the authors could be indeed converted into: the PSII stacks are over-detected in vitro, and it is certainly a simpler explanation for their presence. It is also important to mention that PSII stacking alone is not the only reason for grana architecture - stacking with the antenna of larger complexes, absent in the authors' preparation could also contribute to grana maintenance; and auxiliary proteins such as CURT help with this issue as well. Here a recent demonstration of the importance of minor antenna should probably be also cited: https://doi.org/10.1101/2021.12.31.474624
We used the term “flexible” rather than “transient” to describe the interactions within the stacked PSII dimer. Our data (and tomographic data) do not contain any temporal component. When we used the term under-detected we refer to the fact that PSII is mainly detected by the luminal extrinsic subunits. The flexibility detected in our analysis may affect the concurrent visibly of these features in the PSII complexes making up an individual PSII stack. Specifically, Wietrzynski et al mainly analyze C2S2M2L2 complexes while our analysis only contained C2S2 complexes. It is likely that the different amount of bound LHCII affect PSII stacking as well. For example, Wietrzynski et al, show some overlap between LHCII complexes and little overlap between cores in the larger complexes they analyzed. We observe mainly core to core overlap with little LHCII overlap in the smaller C2S2, although we did not observe any states where LHC’s were not included in what appear to be the binding interface. We agree with the reviewer on the relevance Lhcb’s and CURT contributions to stacking but prefer to focus on what was directly demonstrated in our data. We clearly note that we are discussing in-vitro results.
Taking these last thoughts, I would like to finish by mentioning one more thing - almost philosophical. The authors are certainly at the forefront of the booming cryoEM revolution in biology which is profoundly changing the way we understand the living. There is absolutely zero doubt that this powerful technique is of the highest interest. But a growing number of structures of photosynthetic complexes remain puzzling, in particular with regard to their abundance in vivo (such as the PSII stacks) and functional relevance. How do we ascertain that these interactions are not due to in vitro preparation (isolation from cells, solubilisation)? Which ways can we use to try to exclude this (simple) hypothesis? I suggest that at least a small extent of biological replicas - experiments performed on separate batches, in different technical conditions, with slightly altered solubilization conditions, and so on - could shed light on the nature of these structures and their occurrence in vivo. Technical reps of the freezing+analysis pipeline could also be tried to see the variability. This would strongly reinforce this manuscript and its conclusions, and while not completely unequivocal (the stacked PSII, for example, could form upon each purification), a quantification of the effects would be of high interest.
We certainly share the reviewer hope of being able to conduct cause and effect cryoEM experiments covering a complete set of experimental parameters. This is still beyond reach in terms of time and cost. Within each cryoEM experiment, however, all the analysis is consistent and, more importantly, transparent with regards to image analysis, which is the most important factor in our opinion. Preparation artefacts are always a possibility but, in our opinion, cryoEM is not affected by them differentially compared to other techniques. As we mentioned above, the particles are being observed suspended in vitreous ice, this is not different, and one can say even better, then numerous low temperature spectroscopic observations on samples suspended in glass state or crystals obtained in the presence of high concentrations of various agents. One thing that validates structural studies are the chemical details (bond lengths and angles etc…) underlying every model which are consistence with known values to close tolerances.
Reviewer #3 (Public Review):
In this manuscript, Caspy et al. present a detailed structural analysis of eukaryotic photosystem II (PSII) isolated from the green alga Dunaliella salina. By combining single-particle cryo-EM with multibody refinement, the authors not only reveal a high-resolution (2.4Å) structure of the eukaryotic PSII, but also demonstrate alternate conformations and intrinsic flexibility of the overall complex. Stretched and compact conformations of the PSII dimer were readily identified within the single-particle dataset. From this structural analysis, the authors propose that excitation energy transfer properties may be modulated by changes in transfer distance between key chlorophyll molecules observed in different conformational states of the PSII dimer. Due to the high resolution of the maps obtained, the authors identify post-translational modifications and a sodium binding site based on the observed cryo-EM maps. Additionally, the authors analyze PSII complexes in stacked and unstacked configurations, and find that compact and stretched states also exist within the stacked PSII complexes. From their cryo-EM maps, the authors demonstrate that there is no direct protein-protein interaction between stacked PSII complexes, and rather propose a model wherein long-range electrostatic interactions mediated by divalent cations such as magnesium, can facilitate PSII stacking.
The conclusions and models presented in the manuscript are mostly well justified by the data. The cryo-EM maps are high quality and the models appear generally well refined. However, some aspects of data processing and analysis, as well as the resultant conclusions need to be clarified.
- In general, it is not clear from the cryo-EM processing workflow (suppl. Fig 1) or the methods section when exactly symmetry was applied during 3D classification and refinement. In the case of C2S2 unstacked particles, when was symmetry first applied in the overall processing workflow? To identify the compact and stretched configurations of C2S2, did the 3D classification without alignment (and/or the refinement preceding this classification) have C2 symmetry applied? If so, have you considered the possibility that some particles may actually be asymmetric in some regions?
We modified figure S1 to clearly indicate the use of symmetry and particle expansion. In general, we refined most of the particle sets without symmetry (C1). At the final processing stage of the unstacked PSII sets, after we separated both conformations, we used C2 symmetry to expand the data, this was followed by multibody refinement. No symmetry or symmetry expansion was used for the stacked PSII particle sets.
- Following multibody refinement in Relion individual maps and half-maps for each body will be generated. There is no mention in the methods of how these individual maps for each C2S2 "monomer" were combined to produce an overall map of the dimer following multibody refinement. There are several methods currently used to combine such maps, including taking the maximum or average of the two maps or using a model-based approach in phenix. The authors should be explicit about the method they used, any potential artifacts that may develop from this map combination process, and/or the interface between masks used in multibody refinement.
We used phenix.combined_focused_maps to combine the maps. This is now indicated in the method section.
- In addition to the point raised above, following multibody refinement there will be an individual FSC curve and resolution for each body. However, in supplemental figure 2 and supplemental table 1, only a single FSC curve and resolution are reported. Are these FSC curves/resolutions only reported for the better of the two bodies? If not, how was a single resolution calculated for the overall map of combined bodies?
Both FSC curves were calculated and were highly similar, as expected following C2 expansion. This can also be evaluated from the local resolution maps which are highly similar between the two bodies. The reported resolutions are all taken from the displayed FSC curves generated through relion PostProcess.
- One of the major conclusions from the 3D classification and multibody refinement is that conformational changes and inherent flexibility of the PSII dimers have the potential to change distances between cofactors in the complex, ultimately leading to altered excitation energy transfer. However, it is unclear whether or not the authors believe one conformation over another may more readily support the evolution of oxygen. It would be nice if the authors could elaborate slightly upon this topic in the discussion.
As discussed above the structural changes associated with the formation of quenching centers are not expected to be detected in the current work. The changes we observe can however affect the transfer to such centers and by doing so can play an important part in PSII biology. We do not detect any changes around the OEC and we don’t find any reason to think the two conformations are different with respect to their ETC.
- Along the lines of point 4 above, on line 95 the authors claim that "the high specific activity of 816 umol O2/ (mg Chl * hr) suggest that" both the C2S2 compact and stretched conformation are highly active. However, it is not clear to me why this measure of specific activity would suggest that both PSII conformations should have "high" activity. Maybe a reference here would help guide readers to previous measures of specific activity?
Looking at specific activity from previously published structural studies on eukaryotic PSII we find that Sheng et al, 2019 reported on a specific activity of 272 mol O2/ (mg Chl * hr), this difference can stem partially from the presence of larger complexes in their preparation and is comparable to the activity that we measured in our As fraction (276 mol O2/ (mg Chl * hr), Figure 1-figure supplement 9). Reported specific activity values from plants (Pisum sativum) are also similar, Su et al, reported on a maximal value of 288 mol O2/ (mg Chl * hr), again, for larger complexes which can explain some of the difference. However, the specific activity measured for the C2S2 PSII isolated in the current study is 2.8 X higher than this value, more than the differences in chl content which ranges between 1.5 X to 2 X in favor of the larger complexes. If either one of the conformations is not as active, it would only mean that the other conformation will display even higher specific activity which seems less likely. In addition, we find no difference around the oxygen evolution center or in the peripheral luminal subunits in both the shape or map strength so both orientations show highly similar structures around these regions which determine the oxygen evolution activity.
- It is claimed that "more than 2100 water molecules were detected in the C2S2 compressed model", and the water distribution is shown in Figure 3. Obtaining resolutions capable of visualizing waters with cryo-EM is still a significant challenge. Upon visual inspection of the map supplied, it appears that several of the waters that were built into the atomic model simply do not have supporting peaks in the coulomb potential map above the level of noise. While some of the modeled waters are certainly supported by the map, in my opinion, there are many waters that simply are not, or at best are questionable. What method or tool was originally used to build waters into the model, and how were these waters subsequently validated during structure refinement?
We followed standard methods for water placement and refinement in the preparation of the model, in addition to manually curating the water structure. However, in light of the reviewer comment we undertook additional rounds of refinement and inspection of the water molecules in the model. We removed a few hundred water molecules so that the total number of water molecules is now around 1700. All the water molecules in the present model should be well supported at maps values higher then 2.5 sigma and in our opinion the current water model should be regarded as conservative and underestimates the number of bound water molecules. This also led to some improvements in additional validation statistics of the model which are listed in the Table 1. The new model has been deposited in the PDB and the new PDB validation report is included in our resubmission.
- The authors claim to identify several unique map densities during model building. One of these is a sodium ion close to the OEC, which is coordinated by D1-His337, several backbone carbonyls, and a water molecule. When looking closely at the cryo-EM map supplied, it appears that the coulomb potential map is quite weak for this sodium, and is only visible at quite low contour levels. In fact, the features for the coordinating water, and chloride ions located ~7-9A away are much stronger than the sodium. Do the authors have any explanation for why the cryo-EM map is significantly weaker for the sodium compared to the coordinating water or chloride ions in the same general vicinity? Similar to what they did for the other post-translational modifications, the authors should consider showing the actual cryo-EM map for the bound sodium in supplemental Figure 10 a,b.
Our main support for the placement of a Na+ ion in this location stems from the analysis of Wang et al. Our maps show the presence of a density which is discernible at 4 σ with an elongated shape suggesting the presence of multiple atoms/waters. Although in principle positive ions should have very strong densities in cryoEM maps due to their interactions with electrons, other factors such as occupancy, coordination and b-factor also play a role making the distinction between water and sodium complicated and case specific. The sodium peak is not observed in unsharpened maps (as do most of the water molecules which occupy conserved positions).
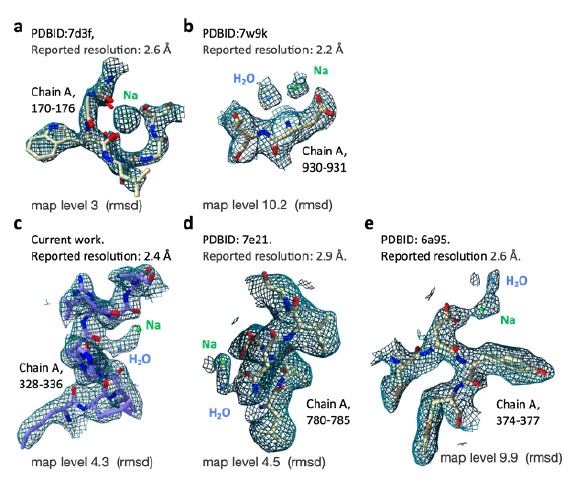 We collected a few examples from comparable cases (cryo-EM maps of similar resolution ranges) where the presence of sodium ions is highly probable based on additional evidence. These maps densities highlight the factors we discussed above. In cases ‘a’ (dual oxidase 1 prepared in high sodium conditions) and ‘b’ (human voltage-gated sodium channel), Na+ is observed in a highly coordinated states and especially in ‘a’ shows the expected increase density values compared to water molecules. However, cases ‘d’ (human Na+/K+ P type Atpase) and ‘e’ (voltage-gated sodium channel) appear very similar to the proposed Na+ assignment in PSII. We conclude that map density alone is not enough to distinguish between Na+ and water molecules and rely on the additional experiments described by Wang et al. which show increase PSII activity in elevated Na+ levels in basic conditions.
We collected a few examples from comparable cases (cryo-EM maps of similar resolution ranges) where the presence of sodium ions is highly probable based on additional evidence. These maps densities highlight the factors we discussed above. In cases ‘a’ (dual oxidase 1 prepared in high sodium conditions) and ‘b’ (human voltage-gated sodium channel), Na+ is observed in a highly coordinated states and especially in ‘a’ shows the expected increase density values compared to water molecules. However, cases ‘d’ (human Na+/K+ P type Atpase) and ‘e’ (voltage-gated sodium channel) appear very similar to the proposed Na+ assignment in PSII. We conclude that map density alone is not enough to distinguish between Na+ and water molecules and rely on the additional experiments described by Wang et al. which show increase PSII activity in elevated Na+ levels in basic conditions.- The cryo-EM maps showing CP29-Ser84 phosphorylation and CP47-Cys218 sulfinylation are quite convincing. However, it is interesting that these modifications are only observed in the compact conformation, and not in the stretched conformation. Can the authors elaborate on whether or not they believe the compact and stretched conformations could be a result of these posttranslational modifications, or vice versa?
This is an interesting suggestion. In our opinion it is less likely that the modification themselves trigger the transition between compact and stretched states. It is not clear how these modifications will stabilize the compact vs the stretched states. It is equally likely that these modifications are somehow triggered by the structural change. We cannot be certain that these modifications are not present in the stretched orientation as well but remain unobserved due to resolution differences. The correlation between the states and post translation modifications should be verified before a discussion on their possible roles in the transitions.
- Do the authors believe that PSII dimers in the solution can readily interconvert between compact and stretched conformations? Or is the relative ratio of these conformations fixed at the time of membrane solubilization with decyl-maltoside?
We think that its more probable that the transition between these states occur in the membrane phase. The main reason for this will be that pigment loss and structural transitions in CP29 are more likely to occur in the membrane rather than in aqueous/micelle environments.
- The model proposed for divalent cation-mediated stacking of PSII dimers is compelling, and seems to be in agreement with previous investigations that observed a lack of stacked dimers in cryo-EM preparations lacking calcium/magnesium. However, my understanding from reading the methods section is that the observed lack of density between the stacked PSII dimers was inferred from maps obtained after multibody refinement. Based on the way the masks to define bodies were created for multibody refinement (Fig. 4A), the region between stacked dimers would be highly prone to map artifacts following multibody refinement. Have the authors looked closely at the interfacial region between stacked dimers following conventional 3D classification/refinement to ensure that there are indeed no features observed in the interfacial region even at low contour levels?
We’ve made several attempts to resolve differences in the space between the stacked PSII dimer. These include focused classification with masks containing selected volumes from this regions and masks that include only one of the stacked PSII dimers to avoid signal subtraction in this region. All of these did not reveal any discernible features in this region. In addition, any stable binding of a bridging protein across the stacked dimer will probably be at least partially visible as additional density over the unstacked PSII. We searched for such features and found none.
-

eLife assessment
The work of Caspy and coworkers resolves the cryo-EM structures of stacked and unstacked PSII supercomplexes of Dunaliella, revealing unexpected connectivity and conformational flexibility, with intriguing implications for the function and regulation of photosynthesis.
-

Reviewer #1 (Public Review):
By careful analyses of single particle images, the authors could identify two distinct structural forms of PSII, a stretched and a compact one. Furthermore, they could analyze a pair of two PSII complexes facing each other on their flat stroma-facing surfaces forming a protein sandwich, which can be made of both the stretched as well as the compact PSII forms. A crucial change in the positioning of the CP29 subunit in the two PSII configurations was identified that favors energy transfer between chlorophylls either to light-harvesting complex II or to the PSII-core antenna complexes. Further aspects like water channels to the oxygen-evolving complex, identification of a Na+ ion, and post-translational modifications were discovered. Overall, the manuscript provides a deep view into the structure of PSII …
Reviewer #1 (Public Review):
By careful analyses of single particle images, the authors could identify two distinct structural forms of PSII, a stretched and a compact one. Furthermore, they could analyze a pair of two PSII complexes facing each other on their flat stroma-facing surfaces forming a protein sandwich, which can be made of both the stretched as well as the compact PSII forms. A crucial change in the positioning of the CP29 subunit in the two PSII configurations was identified that favors energy transfer between chlorophylls either to light-harvesting complex II or to the PSII-core antenna complexes. Further aspects like water channels to the oxygen-evolving complex, identification of a Na+ ion, and post-translational modifications were discovered. Overall, the manuscript provides a deep view into the structure of PSII unraveling novel aspects of its structural flexibility and fine details. The experiments and data analysis were conducted very carefully and thoroughly. The work could spark new discussions about the importance of PSII flexibility in photosynthetic membranes.
-

Reviewer #2 (Public Review):
I am not a specialist in cryo-EM, so cannot comment on the technicalities of the structure reconstruction or methods used. I thus focus on the conclusions and observations that the authors provide in the manuscript and their relevance to functional photosynthesis.
The authors attempt to resolve the structure of PSII from Dunaliella and noticed that three types of PSII could be identified: two conformational states, and a stacked configuration. There is no doubt that these structures add to our current knowledge of PSII and that they exist in abundance upon solubilisation of the sample. My main issue however is the relevance to in vivo conditions, and the efforts to exclude the possibility that pigment loss and conformational states and stacking are a reflection of ex-vivo manipulations.
I see a number of …
Reviewer #2 (Public Review):
I am not a specialist in cryo-EM, so cannot comment on the technicalities of the structure reconstruction or methods used. I thus focus on the conclusions and observations that the authors provide in the manuscript and their relevance to functional photosynthesis.
The authors attempt to resolve the structure of PSII from Dunaliella and noticed that three types of PSII could be identified: two conformational states, and a stacked configuration. There is no doubt that these structures add to our current knowledge of PSII and that they exist in abundance upon solubilisation of the sample. My main issue however is the relevance to in vivo conditions, and the efforts to exclude the possibility that pigment loss and conformational states and stacking are a reflection of ex-vivo manipulations.
I see a number of questions pertaining to this work. Starting from the two conformations of PSII, compact and stretched, the authors say that both are highly active based on oxygen measurements at a saturating light intensity. In the meantime, they report large variations in the chl content and positions of the chlorophyll molecules in these structures (also compared to other known PSIIs). This gives the impression that one can lose two chlorophylls, and freely modify the distance between others without losing efficiency, certainly a risky conclusion. Are the samples highly active also in light-limiting conditions? It is thought that even tiny movements and alterations in chl-chl distances alter their coupling and spectral properties, how come the variations in this report are so huge? In other words, the assay tests the charge separation activity of the PSII RC in the preps, but not the light-harvesting efficiency.
How does one ascertain that the lost chlorophyll molecules in CP29 are not a preparation error? Does slightly increasing the detergent concentration impact the proportion of stretched:compact forms?
On a similar note, how do the authors exclude that a certain interaction with this type of grid impacts the distribution of these complexes? Is it identical to a biologically separate preparation of algae? In case of discoveries of this type, it is of high importance to exclude as many possibilities of non-native conditions or influences on the structure.I would further like to encourage the authors to elaborate on the CP29 phosphorylation. What is the proportion of PSIIcomp that are phosphorylated? I assume it is not 100%, as in this case, the authors would propose that this is the effect that modulates between compact and stretched architectures.
In line 290, the authors highlight the structural heterogeneity within the two groups' PSII conformations. I would like to see how does the distribution look like for all the structures together: are the two (stretched and compact) specifically forming two heterogenous distributions? Or is it possible that the distribution between the two is quasi-continuous? In other words, if the structures are not perfectly defined, how do the authors decide that two- and not more or less subtypes exist?
Considering the stacked PSII, I also have a few concerns. Contrary to previous studies the authors do not assign a functional role to the stacking beyond the structural aspect. This could be better backed by a discussion about the closest chlorophyll a molecules across the stacked PSII, which given the rather large distance shown in fig. 4L seems to be too large for any EET across the stromal gap.
There is a report that suggests the presence of some density between the stacked PSII - could the authors comment on the differences between it and their work? Are the angles and positions conserved between these types of stacks? https://doi.org/10.1038/s41598-017-10700-8Line 387, the authors state that due to the transient nature of the interactions across the stromal gap, the stacks could be "under-detected" in cryo-ET data. This statement is in my opinion misformulated. For once, the transient interaction argument would apply the same (if not more due to changing conditions induced by the purification process) to the single particle analysis performed in this paper. Second, tomographic volumes detect hundreds of PSII in a suspended state. Any transient interaction that adds up to 25% of particle population in a steady state cell should be clearly visible, while the in situ data suggests not more than random cross-stromal-gap orientations. Of course, this can be a specificity of Chlamydomonas or a particular growth condition. The statement used by the authors could be indeed converted into: the PSII stacks are *over-detected in vitro*, and it is certainly a simpler explanation for their presence. It is also important to mention that PSII stacking alone is not the only reason for grana architecture - stacking with the antenna of larger complexes, absent in the authors' preparation could also contribute to grana maintenance; and auxiliary proteins such as CURT help with this issue as well. Here a recent demonstration of the importance of minor antenna should probably be also cited: https://doi.org/10.1101/2021.12.31.474624
Taking these last thoughts, I would like to finish by mentioning one more thing - almost philosophical. The authors are certainly at the forefront of the booming cryoEM revolution in biology which is profoundly changing the way we understand the living. There is absolutely zero doubt that this powerful technique is of the highest interest. But a growing number of structures of photosynthetic complexes remain puzzling, in particular with regard to their abundance in vivo (such as the PSII stacks) and functional relevance. How do we ascertain that these interactions are not due to in vitro preparation (isolation from cells, solubilisation)? Which ways can we use to try to exclude this (simple) hypothesis? I suggest that at least a small extent of biological replicas - experiments performed on separate batches, in different technical conditions, with slightly altered solubilization conditions, and so on - could shed light on the nature of these structures and their occurrence in vivo. Technical reps of the freezing+analysis pipeline could also be tried to see the variability. This would strongly reinforce this manuscript and its conclusions, and while not completely unequivocal (the stacked PSII, for example, could form upon each purification), a quantification of the effects would be of high interest.
-

Reviewer #3 (Public Review):
In this manuscript, Caspy et al. present a detailed structural analysis of eukaryotic photosystem II (PSII) isolated from the green alga Dunaliella salina. By combining single-particle cryo-EM with multibody refinement, the authors not only reveal a high-resolution (2.4Å) structure of the eukaryotic PSII, but also demonstrate alternate conformations and intrinsic flexibility of the overall complex. Stretched and compact conformations of the PSII dimer were readily identified within the single-particle dataset. From this structural analysis, the authors propose that excitation energy transfer properties may be modulated by changes in transfer distance between key chlorophyll molecules observed in different conformational states of the PSII dimer. Due to the high resolution of the maps obtained, the authors …
Reviewer #3 (Public Review):
In this manuscript, Caspy et al. present a detailed structural analysis of eukaryotic photosystem II (PSII) isolated from the green alga Dunaliella salina. By combining single-particle cryo-EM with multibody refinement, the authors not only reveal a high-resolution (2.4Å) structure of the eukaryotic PSII, but also demonstrate alternate conformations and intrinsic flexibility of the overall complex. Stretched and compact conformations of the PSII dimer were readily identified within the single-particle dataset. From this structural analysis, the authors propose that excitation energy transfer properties may be modulated by changes in transfer distance between key chlorophyll molecules observed in different conformational states of the PSII dimer. Due to the high resolution of the maps obtained, the authors identify post-translational modifications and a sodium binding site based on the observed cryo-EM maps. Additionally, the authors analyze PSII complexes in stacked and unstacked configurations, and find that compact and stretched states also exist within the stacked PSII complexes. From their cryo-EM maps, the authors demonstrate that there is no direct protein-protein interaction between stacked PSII complexes, and rather propose a model wherein long-range electrostatic interactions mediated by divalent cations such as magnesium, can facilitate PSII stacking.
The conclusions and models presented in the manuscript are mostly well justified by the data. The cryo-EM maps are high quality and the models appear generally well refined. However, some aspects of data processing and analysis, as well as the resultant conclusions need to be clarified.
1. In general, it is not clear from the cryo-EM processing workflow (suppl. Fig 1) or the methods section when exactly symmetry was applied during 3D classification and refinement. In the case of C2S2 unstacked particles, when was symmetry first applied in the overall processing workflow? To identify the compact and stretched configurations of C2S2, did the 3D classification without alignment (and/or the refinement preceding this classification) have C2 symmetry applied? If so, have you considered the possibility that some particles may actually be asymmetric in some regions?
2. Following multibody refinement in Relion individual maps and half-maps for each body will be generated. There is no mention in the methods of how these individual maps for each C2S2 "monomer" were combined to produce an overall map of the dimer following multibody refinement. There are several methods currently used to combine such maps, including taking the maximum or average of the two maps or using a model-based approach in phenix. The authors should be explicit about the method they used, any potential artifacts that may develop from this map combination process, and/or the interface between masks used in multibody refinement.
3. In addition to the point raised above, following multibody refinement there will be an individual FSC curve and resolution for each body. However, in supplemental figure 2 and supplemental table 1, only a single FSC curve and resolution are reported. Are these FSC curves/resolutions only reported for the better of the two bodies? If not, how was a single resolution calculated for the overall map of combined bodies?
4. One of the major conclusions from the 3D classification and multibody refinement is that conformational changes and inherent flexibility of the PSII dimers have the potential to change distances between cofactors in the complex, ultimately leading to altered excitation energy transfer. However, it is unclear whether or not the authors believe one conformation over another may more readily support the evolution of oxygen. It would be nice if the authors could elaborate slightly upon this topic in the discussion.
5. Along the lines of point 4 above, on line 95 the authors claim that "the high specific activity of 816 umol O2/ (mg Chl * hr) suggest that" both the C2S2 compact and stretched conformation are highly active. However, it is not clear to me why this measure of specific activity would suggest that both PSII conformations should have "high" activity. Maybe a reference here would help guide readers to previous measures of specific activity?
6. It is claimed that "more than 2100 water molecules were detected in the C2S2 compressed model", and the water distribution is shown in Figure 3. Obtaining resolutions capable of visualizing waters with cryo-EM is still a significant challenge. Upon visual inspection of the map supplied, it appears that several of the waters that were built into the atomic model simply do not have supporting peaks in the coulomb potential map above the level of noise. While some of the modeled waters are certainly supported by the map, in my opinion, there are many waters that simply are not, or at best are questionable. What method or tool was originally used to build waters into the model, and how were these waters subsequently validated during structure refinement?
7. The authors claim to identify several unique map densities during model building. One of these is a sodium ion close to the OEC, which is coordinated by D1-His337, several backbone carbonyls, and a water molecule. When looking closely at the cryo-EM map supplied, it appears that the coulomb potential map is quite weak for this sodium, and is only visible at quite low contour levels. In fact, the features for the coordinating water, and chloride ions located ~7-9A away are much stronger than the sodium. Do the authors have any explanation for why the cryo-EM map is significantly weaker for the sodium compared to the coordinating water or chloride ions in the same general vicinity? Similar to what they did for the other post-translational modifications, the authors should consider showing the actual cryo-EM map for the bound sodium in supplemental Figure 10 a,b.
8. The cryo-EM maps showing CP29-Ser84 phosphorylation and CP47-Cys218 sulfinylation are quite convincing. However, it is interesting that these modifications are only observed in the compact conformation, and not in the stretched conformation. Can the authors elaborate on whether or not they believe the compact and stretched conformations could be a result of these posttranslational modifications, or vice versa?
9. Do the authors believe that PSII dimers in the solution can readily interconvert between compact and stretched conformations? Or is the relative ratio of these conformations fixed at the time of membrane solubilization with decyl-maltoside?
10. The model proposed for divalent cation-mediated stacking of PSII dimers is compelling, and seems to be in agreement with previous investigations that observed a lack of stacked dimers in cryo-EM preparations lacking calcium/magnesium. However, my understanding from reading the methods section is that the observed lack of density between the stacked PSII dimers was inferred from maps obtained after multibody refinement. Based on the way the masks to define bodies were created for multibody refinement (Fig. 4A), the region between stacked dimers would be highly prone to map artifacts following multibody refinement. Have the authors looked closely at the interfacial region between stacked dimers following conventional 3D classification/refinement to ensure that there are indeed no features observed in the interfacial region even at low contour levels?
-