Modelling the response to vaccine in non-human primates to define SARS-CoV-2 mechanistic correlates of protection
Curation statements for this article:-
Curated by eLife
Evaluation Summary:
This work should be of interest to a broad readership in infectious diseases, especially those people interested in modeling of infections. It combines statistical and mechanistic modeling to find assayable correlates of immunity for vaccines. This method could be relevant to many diseases or vaccines, although the particular markers identified here likely will be limited in their generalizability.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. The reviewers remained anonymous to the authors.)
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
- Evaluated articles (ScreenIT)
Abstract
The definition of correlates of protection is critical for the development of next-generation SARS-CoV-2 vaccine platforms. Here, we propose a model-based approach for identifying mechanistic correlates of protection based on mathematical modelling of viral dynamics and data mining of immunological markers. The application to three different studies in non-human primates evaluating SARS-CoV-2 vaccines based on CD40-targeting, two-component spike nanoparticle and mRNA 1273 identifies and quantifies two main mechanisms that are a decrease of rate of cell infection and an increase in clearance of infected cells. Inhibition of RBD binding to ACE2 appears to be a robust mechanistic correlate of protection across the three vaccine platforms although not capturing the whole biological vaccine effect. The model shows that RBD/ACE2 binding inhibition represents a strong mechanism of protection which required significant reduction in blocking potency to effectively compromise the control of viral replication.
Article activity feed
-

-

Author Responses
Reviewer #3 (Public Review):
Alexandre et al. fit a mathematical model of viral-host dynamics to previously-published data from three SARS-CoV-2 challenge studies in non-human primates and identify immune markers that correlate with "protection" (as measured by viral loads) as well or better than knowing whether an animal was naive, vaccinated, or recovered from natural infection. Crucially, the use of this model allowed for summarizing the complex time-dependent outcome data (viral sgRNA and gRNA loads over time) as a small number of more interpretable parameters (e.g., within-host viral infectivity, infected cell death rates, virion production rates) while allowing for intra-individual variation in a statistically rigorous fashion. Vaccine correlates of protection are notoriously difficult to identify and could be …
Author Responses
Reviewer #3 (Public Review):
Alexandre et al. fit a mathematical model of viral-host dynamics to previously-published data from three SARS-CoV-2 challenge studies in non-human primates and identify immune markers that correlate with "protection" (as measured by viral loads) as well or better than knowing whether an animal was naive, vaccinated, or recovered from natural infection. Crucially, the use of this model allowed for summarizing the complex time-dependent outcome data (viral sgRNA and gRNA loads over time) as a small number of more interpretable parameters (e.g., within-host viral infectivity, infected cell death rates, virion production rates) while allowing for intra-individual variation in a statistically rigorous fashion. Vaccine correlates of protection are notoriously difficult to identify and could be extremely valuable when assessing risks and designing vaccine dosages and booster schedules. The methodological approach developed in this paper is broadly applicable and a worth-while contribution by itself. In the context of the particular data analyzed here, the statistically-predictive immune markers showed reassuring consistency between the two studies using protein-based vaccines, although the third study using a mRNA-based vaccine differed. The conclusions have two limitations, the first of which is directly acknowledged by the authors while the second is not:
- The definition of "protection" is limited to the within-host cellular level. While within-host transmission is certainly related to between-host transmission and disease severity, many other factors play a role as well; this limitation is nicely acknowledged by the authors.
- The models may be overfit to the data, although this concern is somewhat tempered by the finding that application to the two protein-based vaccine studies yielded broadly similar results. Predictive statistical models of the type used here would ideally be tested on a held-out set of test data from the same type of experiment. The repeated use of BIC in a stepwise model selection framework with many predictors and limited biological replicates is risky.
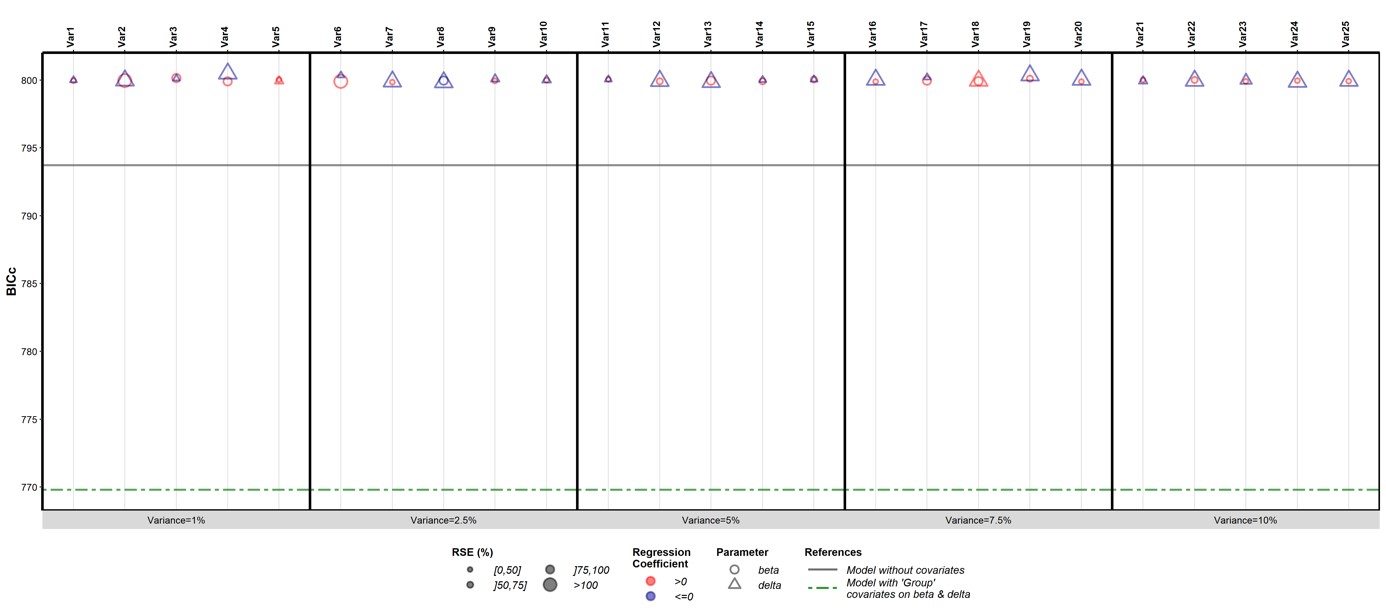
To address the reviewer’s comment about the repeated use of BIC as a model selection criterion in a stepwise selection procedure, we performed a small simulation to ensure the robustness of BIC despite the multiplicity of tests. We simulated, for each of the 18 NHPs, 25 longitudinal variables as white-noise random variables by varying the variances from 1 to 10%. Figure 1 shows the results we obtained after applying our algorithm with these variables as time-varying covariates. In the figure, the vertical black solid line represents the value of BIC obtained with the model without covariates, and the green dashed line the one obtained with β and δ adjusted for groups. Results appears as robust to the multiplicity of the tests as all adjustments for white-noise variables yield similar BIC values and degrade the model, compared to the one without covariates.
In addition, as mentioned in our response to the comment 4b) of the reviewer 1, we tested the robustness of the results using several selection criteria (AIC, BIC, LL, interindividual variability). All criteria led to similar results.
To mention this point in the manuscript, we created the Appendix 2 “BICc as selection criteria and multiple testing adjustment” in which we present this additional work. This additional file was mentioned in the manuscript at the page 31, Line 666.
-

Evaluation Summary:
This work should be of interest to a broad readership in infectious diseases, especially those people interested in modeling of infections. It combines statistical and mechanistic modeling to find assayable correlates of immunity for vaccines. This method could be relevant to many diseases or vaccines, although the particular markers identified here likely will be limited in their generalizability.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. The reviewers remained anonymous to the authors.)
-

Reviewer #1 (Public Review):
In this study, the authors formulate a model of within-host viral infection (similar to other standard published models) to analyze data of an experiment of a SARS-CoV-2 vaccine in macaques. For this experiment, they have frequent viral load measurements, as well as multiple (>40) other immune parameter measured at multiple time points. The objective of the paper, and the main novelty, is to understand the mechanistic correlates of protection afforded by the vaccine. To this end, the authors fit the mechanistic model to the viral load data in the vaccinated and control macaques (including previously infected) to find what parameters of the model are statistically different among the different experimental groups (this has been done before). But then go a step further and assess which of the more than 40 …
Reviewer #1 (Public Review):
In this study, the authors formulate a model of within-host viral infection (similar to other standard published models) to analyze data of an experiment of a SARS-CoV-2 vaccine in macaques. For this experiment, they have frequent viral load measurements, as well as multiple (>40) other immune parameter measured at multiple time points. The objective of the paper, and the main novelty, is to understand the mechanistic correlates of protection afforded by the vaccine. To this end, the authors fit the mechanistic model to the viral load data in the vaccinated and control macaques (including previously infected) to find what parameters of the model are statistically different among the different experimental groups (this has been done before). But then go a step further and assess which of the more than 40 immune measurements better explains the statistical differences. That is, the inclusion of that information in model should explain the statistical differences in parameters. This is a clever and elegant approach and allows new insights into the mechanisms elicited by the vaccine that help control the virus.
The main strengths of the paper are the wealth of data and the good use of it, with sound statistical and modeling methodology. It is also important that the authors analyze more than one type of vaccine experiments, demonstrating some robustness of the approach. The results obtained, although not totally unexpected, shed some light into the mechanism of action of the vaccine and what experimental assay could serve as a correlate of protection also for other vaccines.
The main weakness of the paper is that currently, the paper is difficult to follow in some parts, and more/ clearer details would benefit the reader. For example, some modeling choices are not properly justified.
-

Reviewer #2 (Public Review):
The manuscript is a body of work focused on defining a "correlate of protection" for SARS-CoV-2 antibody-based vaccines. Their main conclusion, which was the antibody binding was most correlative, is in line with other studies indicating that the vaccines-induced antibodies are protective. The application of a mathematical model of viral infection helped quantify the data, and agreed that the antibodies inhibit the infection of cells. The authors claim that they established a "novel mathematical framework" to define correlates of protection, but the methods were not particularly new and lacked assessment of important vaccine-induced immune responses that would also be considered protective. In addition, it's unclear why one would desire a clinically-relevant correlate of protection without description of how …
Reviewer #2 (Public Review):
The manuscript is a body of work focused on defining a "correlate of protection" for SARS-CoV-2 antibody-based vaccines. Their main conclusion, which was the antibody binding was most correlative, is in line with other studies indicating that the vaccines-induced antibodies are protective. The application of a mathematical model of viral infection helped quantify the data, and agreed that the antibodies inhibit the infection of cells. The authors claim that they established a "novel mathematical framework" to define correlates of protection, but the methods were not particularly new and lacked assessment of important vaccine-induced immune responses that would also be considered protective. In addition, it's unclear why one would desire a clinically-relevant correlate of protection without description of how to measure it in the clinic with a defining level of how to quantify risk.
-

Reviewer #3 (Public Review):
Alexandre et al. fit a mathematical model of viral-host dynamics to previously-published data from three SARS-CoV-2 challenge studies in non-human primates and identify immune markers that correlate with "protection" (as measured by viral loads) as well or better than knowing whether an animal was naive, vaccinated, or recovered from natural infection. Crucially, the use of this model allowed for summarizing the complex time-dependent outcome data (viral sgRNA and gRNA loads over time) as a small number of more interpretable parameters (e.g., within-host viral infectivity, infected cell death rates, virion production rates) while allowing for intra-individual variation in a statistically rigorous fashion. Vaccine correlates of protection are notoriously difficult to identify and could be extremely valuable …
Reviewer #3 (Public Review):
Alexandre et al. fit a mathematical model of viral-host dynamics to previously-published data from three SARS-CoV-2 challenge studies in non-human primates and identify immune markers that correlate with "protection" (as measured by viral loads) as well or better than knowing whether an animal was naive, vaccinated, or recovered from natural infection. Crucially, the use of this model allowed for summarizing the complex time-dependent outcome data (viral sgRNA and gRNA loads over time) as a small number of more interpretable parameters (e.g., within-host viral infectivity, infected cell death rates, virion production rates) while allowing for intra-individual variation in a statistically rigorous fashion. Vaccine correlates of protection are notoriously difficult to identify and could be extremely valuable when assessing risks and designing vaccine dosages and booster schedules. The methodological approach developed in this paper is broadly applicable and a worth-while contribution by itself. In the context of the particular data analyzed here, the statistically-predictive immune markers showed reassuring consistency between the two studies using protein-based vaccines, although the third study using a mRNA-based vaccine differed. The conclusions have two limitations, the first of which is directly acknowledged by the authors while the second is not:
1. The definition of "protection" is limited to the within-host cellular level. While within-host transmission is certainly related to between-host transmission and disease severity, many other factors play a role as well; this limitation is nicely acknowledged by the authors.
2. The models may be overfit to the data, although this concern is somewhat tempered by the finding that application to the two protein-based vaccine studies yielded broadly similar results. Predictive statistical models of the type used here would ideally be tested on a held-out set of test data from the same type of experiment. The repeated use of BIC in a stepwise model selection framework with many predictors and limited biological replicates is risky.
-

SciScore for 10.1101/2021.10.29.466418: (What is this?)
Please note, not all rigor criteria are appropriate for all manuscripts.
Table 1: Rigor
NIH rigor criteria are not applicable to paper type.Table 2: Resources
Antibodies Sentences Resources Evaluation of anti-Spike, anti-RBD and neutralizing IgG antibodies: Anti-Spike IgG were titrated by multiplex bead assay: Briefly, Luminex beads were coupled to the Spike protein as previously described (63) and added to a Bio-Plex plate (BioRad). anti-Spike, anti-RBD and neutralizing IgGsuggested: NoneAnti-Spike IgGsuggested: NoneBeads were then washed and anti-NHP IgG-PE secondary antibody (Southern Biotech, clone SB108a) was added at a 1:500 dilution for 45 min at room temperature. anti-NHP IgG-PEsuggested: NoneAfter incubation, detection antibody was added (MSD SULFO-TAGTM Anti-Human IgG Antibody) … SciScore for 10.1101/2021.10.29.466418: (What is this?)
Please note, not all rigor criteria are appropriate for all manuscripts.
Table 1: Rigor
NIH rigor criteria are not applicable to paper type.Table 2: Resources
Antibodies Sentences Resources Evaluation of anti-Spike, anti-RBD and neutralizing IgG antibodies: Anti-Spike IgG were titrated by multiplex bead assay: Briefly, Luminex beads were coupled to the Spike protein as previously described (63) and added to a Bio-Plex plate (BioRad). anti-Spike, anti-RBD and neutralizing IgGsuggested: NoneAnti-Spike IgGsuggested: NoneBeads were then washed and anti-NHP IgG-PE secondary antibody (Southern Biotech, clone SB108a) was added at a 1:500 dilution for 45 min at room temperature. anti-NHP IgG-PEsuggested: NoneAfter incubation, detection antibody was added (MSD SULFO-TAGTM Anti-Human IgG Antibody) and then MSD GOLDTM Read Buffer B was added and plates read using a MESO QuickPlex SQ 120MM Reader. Anti-Human IgGsuggested: NoneAntibody measurements after the second exposure to SARS-CoV-2. Fig. S5. S5suggested: NoneExperimental Models: Cell Lines Sentences Resources The Nucleocapsid and the Spike RBD domain (Genbank # NC_045512.2) were cloned and produced in E. Coli and CHO cells, respectively, as previously described (31). CHOsuggested: CLS Cat# 603479/p746_CHO, RRID:CVCL_0213)Software and Algorithms Sentences Resources The EC50 value of each sample was determined using GraphPad Prism 8 and antibody titer was calculated as log (1/EC50). GraphPad Prismsuggested: (GraphPad Prism, RRID:SCR_002798)Model parameters were estimated with the SAEM algorithm (Monolix® software version 2019R1). Monolix®suggested: NoneGraphs were generated using R version 3.6.1 and Excel 2016 and details on the statistical analysis for the experiments can be found in the accompanying figure legends. Excelsuggested: NoneResults from OddPub: Thank you for sharing your code.
Results from LimitationRecognizer: We detected the following sentences addressing limitations in the study:One limitation of our study is that the prediction potential of our model relies on the range of the immune markers measured. However, our approach would allow a full exploitation of the data generated as in systems serology where non-neutralizing Ab functions, such as Ab-dependent cellular cytotoxicity (ADCC), Ab-dependent cellular phagocytosis (ADCP), Ab-dependent complement deposition (ADCD), and Ab-dependent respiratory burst (ADRB) are explored (38). The role of ADCC in natural infection has been previously shown (39), ADCD in DNA vaccine recipients (11) and with Ad26 vaccine (40). Here, we extended significantly these data by modelling the viral dynamic, showing that two other protein-based vaccines exert an additional effect on infected cell death which relied on the level of IgG anti-RBD binding antibodies especially for the CD40.RBD targeting vaccine. Measurements of other non-neutralizing Ab functions would probably also capture this additional effect. The next question after determining which marker is a valid mCoP is to define the concentration that leads to protection, looking for a threshold effect that will help to define an objective (10, 41). In the context of SARS-CoV-2 virus, several emerged variants are leading to a significant reduction of viral neutralization as measured by various approaches. However, a 20-fold reduction of viral neutralization might not translate in 20-fold reduction of vaccine efficacy (42). First, there are many steps between viral n...
Results from TrialIdentifier: No clinical trial numbers were referenced.
Results from Barzooka: We did not find any issues relating to the usage of bar graphs.
Results from JetFighter: We did not find any issues relating to colormaps.
Results from rtransparent:- Thank you for including a conflict of interest statement. Authors are encouraged to include this statement when submitting to a journal.
- Thank you for including a funding statement. Authors are encouraged to include this statement when submitting to a journal.
- No protocol registration statement was detected.
Results from scite Reference Check: We found no unreliable references.
-