Conserved structural elements specialize ATAD1 as a membrane protein extraction machine
Curation statements for this article:-
Curated by eLife
Evaluation Summary:
This work extends our understanding of the ATAD1 family of AAA proteins responsible for extracting tail-anchored (TA) proteins mistargeted to the mitochondria. The conclusions of this work are largely consistent to prior structural studies from the same group, but provide clarifications of specific details that will be of interest to those working on these important proteins.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #3 agreed to share their name with the authors.)
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
The mitochondrial AAA ( A TPase A ssociated with diverse cellular A ctivities) protein ATAD1 (in humans; Msp1 in yeast) removes mislocalized membrane proteins, as well as stuck import substrates from the mitochondrial outer membrane, facilitating their re-insertion into their cognate organelles and maintaining mitochondria’s protein import capacity. In doing so, it helps to maintain proteostasis in mitochondria. How ATAD1 tackles the energetic challenge to extract hydrophobic membrane proteins from the lipid bilayer and what structural features adapt ATAD1 for its particular function has remained a mystery. Previously, we determined the structure of Msp1 in complex with a peptide substrate (Wang et al., 2020). The structure showed that Msp1’s mechanism follows the general principle established for AAA proteins while adopting several structural features that specialize it for its function. Among these features in Msp1 was the utilization of multiple aromatic amino acids to firmly grip the substrate in the central pore. However, it was not clear whether the aromatic nature of these amino acids were required, or if they could be functionally replaced by aliphatic amino acids. In this work, we determined the cryo-EM structures of the human ATAD1 in complex with a peptide substrate at near atomic resolution. The structures show that phylogenetically conserved structural elements adapt ATAD1 for its function while generally adopting a conserved mechanism shared by many AAA proteins. We developed a microscopy-based assay reporting on protein mislocalization, with which we directly assessed ATAD1’s activity in live cells and showed that both aromatic amino acids in pore-loop 1 are required for ATAD1’s function and cannot be substituted by aliphatic amino acids. A short α-helix at the C-terminus strongly facilitates ATAD1’s oligomerization, a structural feature that distinguishes ATAD1 from its closely related proteins.
Article activity feed
-

Author Response*
Reviewer #3 (Public Review):
AAA protein are involved in a variety of cellular activity. They all share the same structural fold and still they are all incredibly specialised. This study works towards the direction of understanding the unique specialisation of the AAA protein ATAD1. While the general mechanism of substrate threading by AAA proteins is by now fairly well-elucidated, it remains to describe and understand the finer structural protein details that make each specific AAA perform unfolding (threading) of certain substrate rather than others. Additionally, regulation and stabilisation of each AAA is also finely regulated by specific subdomain.
This work is definitively strong in addressing these two points for ATAD1.
The structural data are solid and the analysis of the pore loops residues and the role …
Author Response*
Reviewer #3 (Public Review):
AAA protein are involved in a variety of cellular activity. They all share the same structural fold and still they are all incredibly specialised. This study works towards the direction of understanding the unique specialisation of the AAA protein ATAD1. While the general mechanism of substrate threading by AAA proteins is by now fairly well-elucidated, it remains to describe and understand the finer structural protein details that make each specific AAA perform unfolding (threading) of certain substrate rather than others. Additionally, regulation and stabilisation of each AAA is also finely regulated by specific subdomain.
This work is definitively strong in addressing these two points for ATAD1.
The structural data are solid and the analysis of the pore loops residues and the role of a11 overall convincing.
- The cell fluorescence microscopy assay is a very good tool for checking in the cell the hypothesis risen by analysing of the structure. However, the assay is currently only based on the localisation of the Gos28 substrate, which leaves open the possibility that ATAD1 a11 mutants will have a different phenotype on different substrates.
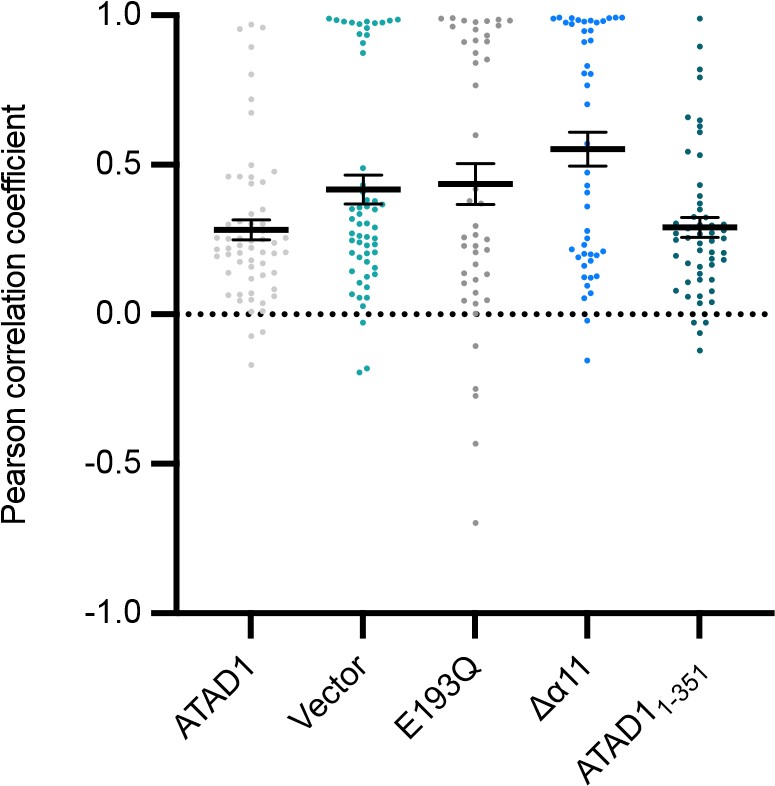
We agree with the reviewer that it would be interesting to test ATAD1’s activity on other known substrates. To do that, we picked Pex26, an established tail-anchored protein substrate of ATAD1. We stably expressed EGFP-Pex26 in ATAD1-/- cells and tested the effect of ATAD1 expression on Pex26 mislocalization. As shown in the figure below, we found that although the general trend observed for Gos28 also holds true for Pex26, the measured PCC values clearly have a bimodal distribution, with some cells showing the complete mislocalization (PCC = 1.0) of Pex26. One exciting possibility to explain this result is that Pex26 is important in peroxisome biogenesis. Once enough Pex26 is mislocalized to the mitochondria, peroxisomal biogenesis becomes impaired, thus causing less Pex26 to be correctly inserted. A partial impairment in Pex26 peroxisomal insertion in turn creates a vicious cycle that leads to the complete mislocalization of Pex26. It will be an interesting to follow up on the cause of this bimodal distribution, which, however, is beyond the scope of this paper.
*Quantification of live-cell imaging showing using the localization of EGFP-Pex26 as a readout. Mean Pearson correlation coefficient (PCC) values and the SEM between EGFP-Pex26 and the mitochondria when expressing the ATAD1 variants indicated. Individual cell PCC values are represented as a single dot. *
-

Evaluation Summary:
This work extends our understanding of the ATAD1 family of AAA proteins responsible for extracting tail-anchored (TA) proteins mistargeted to the mitochondria. The conclusions of this work are largely consistent to prior structural studies from the same group, but provide clarifications of specific details that will be of interest to those working on these important proteins.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #3 agreed to share their name with the authors.)
-

Reviewer #1 (Public Review):
In this work, the authors determine the structure of ATAD1, a AAA protein responsible for removal of mistargeted tail anchored (TA) proteins from the mitochondria. In prior work, this group determined the structure of the yeast ortholog Msp1 and found that aromatic residues in key pore-loops were important for engaging substrates. In the current manuscript, the cryo-EM structure of ATAD1 reveals large similarities with the yeast ortholog but elaborates some details about interactions between subunits and unresolved regions from the prior work. Most important is the presence of an extended helix (a11) nestled between the subunits that was not visible in the prior cryo-EM Msp1 structure but was present in a published crystal structure from another group (Wohlever, et al. 2017).
Based on similar structural …
Reviewer #1 (Public Review):
In this work, the authors determine the structure of ATAD1, a AAA protein responsible for removal of mistargeted tail anchored (TA) proteins from the mitochondria. In prior work, this group determined the structure of the yeast ortholog Msp1 and found that aromatic residues in key pore-loops were important for engaging substrates. In the current manuscript, the cryo-EM structure of ATAD1 reveals large similarities with the yeast ortholog but elaborates some details about interactions between subunits and unresolved regions from the prior work. Most important is the presence of an extended helix (a11) nestled between the subunits that was not visible in the prior cryo-EM Msp1 structure but was present in a published crystal structure from another group (Wohlever, et al. 2017).
Based on similar structural motifs in related AAA proteins, the authors hypothesize that a11 is important for oligomerization and ATAD1 activity. Indeed SEC and activity assays suggest that ADAT1∆a11 assembles poorly, has reduced ATP hydrolysis activity, and fails to bind peptide substrates as readily. Using a novel in vivo mislocalization assay, the authors also show that there are defects with the function of this variant consistent with reduced activity and a failure to form oligomers.
Overall this work extends our understanding of a family of AAA proteins responsible for extracting TA proteins mistargeted to the mitochondria.
-

Reviewer #2 (Public Review):
The AAA+ proteins ATAD1/Msp1 extract mislocalized TA-proteins from the outer membrane of mitochondria allowing for substrate retargeting to the ER. Msp1/ATAD1 belong to the meiotic clade of AAA+ proteins also including katanin and spastin that sever microtubules. The authors previously determined the cryo EM structure of C. thermophilum Msp1, now they report on the structure of the human homolog ATAD1. ATAD1 hexamers were determined two distinct (open vs closed) structures. The main difference between these structures is the position of the seam (M6) subunit, which contacts the clockwise subunit in the closed state. This structure represents an intermediate state in the ATPase and threading cycle of the AAA+ protein.
The authors additionally report on unique structural features that may enable ATAD1 …
Reviewer #2 (Public Review):
The AAA+ proteins ATAD1/Msp1 extract mislocalized TA-proteins from the outer membrane of mitochondria allowing for substrate retargeting to the ER. Msp1/ATAD1 belong to the meiotic clade of AAA+ proteins also including katanin and spastin that sever microtubules. The authors previously determined the cryo EM structure of C. thermophilum Msp1, now they report on the structure of the human homolog ATAD1. ATAD1 hexamers were determined two distinct (open vs closed) structures. The main difference between these structures is the position of the seam (M6) subunit, which contacts the clockwise subunit in the closed state. This structure represents an intermediate state in the ATPase and threading cycle of the AAA+ protein.
The authors additionally report on unique structural features that may enable ATAD1 fulfilling its specific function in membrane extraction of TA-proteins. They show that ATAD1 harbors a particularly long C-terminal a-helix 11. This extension was not visible in the former Ct Msp1 structure. Notably, other meiotic family members harbor a shorter a11 but an additional a12. ATAD1 a11 contacts the counterclockwise subunit in the hexameric AAA ring, implicating a role in hexamer stabilization.
Finally, the authors established a new, microscopic assay to study ATAD1/Msp1 activity in vivo. This assay is based on the direct visualization of mistargeting of the TA-protein GFP-Gos28. By co-expressing the fluorescent reporter and ATAD1 mutants in ATAD1+/+ and ATAD1-/- cells, the authors can differentiate between dominant loss-of-function mutants (toxic in ATAD1+/+ cells) and recessive mutants. This assay proves to be very useful for analysis of ATAD1 activity and allowed documenting oligomerization defects of ATAD1 a11 deletions, which was confirmed in vitro by analysis of respective purified proteins.
-

Reviewer #3 (Public Review):
AAA protein are involved in a variety of cellular activity. They all share the same structural fold and still they are all incredibly specialised. This study works towards the direction of understanding the unique specialisation of the AAA protein ATAD1. While the general mechanism of substrate threading by AAA proteins is by now fairly well-elucidated, it remains to describe and understand the finer structural protein details that make each specific AAA perform unfolding (threading) of certain substrate rather than others. Additionally, regulation and stabilisation of each AAA is also finely regulated by specific subdomain.
This work is definitively strong in addressing these two points for ATAD1.
The structural data are solid and the analysis of the pore loops residues and the role of a11 overall …Reviewer #3 (Public Review):
AAA protein are involved in a variety of cellular activity. They all share the same structural fold and still they are all incredibly specialised. This study works towards the direction of understanding the unique specialisation of the AAA protein ATAD1. While the general mechanism of substrate threading by AAA proteins is by now fairly well-elucidated, it remains to describe and understand the finer structural protein details that make each specific AAA perform unfolding (threading) of certain substrate rather than others. Additionally, regulation and stabilisation of each AAA is also finely regulated by specific subdomain.
This work is definitively strong in addressing these two points for ATAD1.
The structural data are solid and the analysis of the pore loops residues and the role of a11 overall convincing.
The cell fluorescence microscopy assay is a very good tool for checking in the cell the hypothesis risen by analysing of the structure. However, the assay is currently only based on the localisation of the Gos28 substrate, which leaves open the possibility that ATAD1 a11 mutants will have a different phenotype on different substrates.
Overall the work is a solid follow up of the work on Msp1 and advances slowly but soundly the knowledge on ATAD1 and its mechanism in the rescue of mislocalised TA proteins. -

-