Body mass index and childhood symptoms of depression, anxiety, and attention-deficit hyperactivity disorder: A within-family Mendelian randomization study
Curation statements for this article:-
Curated by eLife
Evaluation Summary:
This paper uses a new statistical approach called within family Mendelian randomization and asserts that claims of childhood BMI affecting a range of psychiatric traits are unfounded and were mainly caused by confounders that this new approach is able to better identify and control for. They do find a role for maternal BMI on a child's risk for developing depression. The main issue raised is that they do not convincingly show if they do not replicate the old association of childhood BMI with a range of psychiatric traits due to their technique simply having lower power to detect the signal or due to a true lack of this effect.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #2 agreed to share their name with the authors.)
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Higher BMI in childhood is associated with emotional and behavioural problems, but these associations may not be causal. Results of previous genetic studies imply causal effects but may reflect influence of demography and the family environment.
Methods:
This study used data on 40,949 8-year-old children and their parents from the Norwegian Mother, Father and Child Cohort Study (MoBa) and Medical Birth Registry of Norway (MBRN). We investigated the impact of BMI on symptoms of depression, anxiety, and attention-deficit hyperactivity disorder (ADHD) at age 8. We applied within-family Mendelian randomization, which accounts for familial effects by controlling for parental genotype.
Results:
Within-family Mendelian randomization estimates using genetic variants associated with BMI in adults suggested that a child’s own BMI increased their depressive symptoms (per 5 kg/m 2 increase in BMI, beta = 0.26 S.D., CI = −0.01,0.52, p=0.06) and ADHD symptoms (beta = 0.38 S.D., CI = 0.09,0.63, p=0.009). These estimates also suggested maternal BMI, or related factors, may independently affect a child’s depressive symptoms (per 5 kg/m 2 increase in maternal BMI, beta = 0.11 S.D., CI:0.02,0.09, p=0.01). However, within-family Mendelian randomization using genetic variants associated with retrospectively-reported childhood body size did not support an impact of BMI on these outcomes. There was little evidence from any estimate that the parents’ BMI affected the child’s ADHD symptoms, or that the child’s or parents’ BMI affected the child’s anxiety symptoms.
Conclusions:
We found inconsistent evidence that a child’s BMI affected their depressive and ADHD symptoms, and little evidence that a child’s BMI affected their anxiety symptoms. There was limited evidence of an influence of parents’ BMI. Genetic studies in samples of unrelated individuals, or using genetic variants associated with adult BMI, may have overestimated the causal effects of a child’s own BMI.
Funding:
This research was funded by the Health Foundation. It is part of the HARVEST collaboration, supported by the Research Council of Norway. Individual co-author funding: the European Research Council, the South-Eastern Norway Regional Health Authority, the Research Council of Norway, Helse Vest, the Novo Nordisk Foundation, the University of Bergen, the South-Eastern Norway Regional Health Authority, the Trond Mohn Foundation, the Western Norway Regional Health Authority, the Norwegian Diabetes Association, the UK Medical Research Council. The Medical Research Council (MRC) and the University of Bristol support the MRC Integrative Epidemiology Unit.
Article activity feed
-

-

Author Response
Reviewer #2 (Public Review):
This study evaluates the causal relationship between childhood obesity on the one hand, and childhood emotional and behavioral problems on the other. It applies Mendelian Randomization (MR), a family of methods in statistical genetics that uses genetic markers to break the symmetry between correlated traits, allowing inference of causation rather than mere correlation. The authors argue convincingly that previous studies of these traits, both those using non-genetic observational epidemiology methods and those using standard MR methods, may be confounded by demographic effects and familial effects. One possible example of this kind of confounding is that the idea that obesity in parents may contribute to emotional and behavioral problems in children; another is the idea that adults with …
Author Response
Reviewer #2 (Public Review):
This study evaluates the causal relationship between childhood obesity on the one hand, and childhood emotional and behavioral problems on the other. It applies Mendelian Randomization (MR), a family of methods in statistical genetics that uses genetic markers to break the symmetry between correlated traits, allowing inference of causation rather than mere correlation. The authors argue convincingly that previous studies of these traits, both those using non-genetic observational epidemiology methods and those using standard MR methods, may be confounded by demographic effects and familial effects. One possible example of this kind of confounding is that the idea that obesity in parents may contribute to emotional and behavioral problems in children; another is the idea that adults with emotional and behavioral issues may be more likely to have children with partners who are obese, and vice-versa. They then make use of a recently proposed "within-family" MR method, which should effectively control for these confounders, at the cost of higher uncertainty in the estimated effect size, and therefore lower power to detect small effects. They report that none of the previously reported associations of childhood BMI with anxiety, depression, or ADHD are replicated using the within-family MR method, and that in the case of depression the primary association appears to be with maternal BMI rather than the child's own BMI.
This argument that these confounders may affect these phenotypes is fairly sound, and within-family MR should indeed do a good job of controlling for them. I do not see any major issues with the cohort itself or the choice of genetic instruments. I also do not see any major issues with the definitions or ascertainment of the phenotypes studied, though I am not an expert on any of these phenotypes in particular. I am especially satisfied with the series of analyses demonstrating that the results are robust to many variations of MR methodology. Overall, I think the positive result this study reports is very credible: that the known association between childhood BMI and depression is likely primarily due to an effect of maternal BMI rather than the child's own BMI (though given that paternal BMI has a similar effect size with only a slightly wider confidence interval, I would instead say that the effect is from parental BMI generally, not specifically maternal.)
In the updated results based on the larger genetic data release, the estimates for the association of maternal BMI and paternal BMI with the child’s depressive symptoms are more clearly different than they were in the smaller dataset (for maternal BMI, beta= 0.11, CI:0.02,0.19, p=0.01; for paternal BMI, beta=0.02, CI:-0.09,0.12, p=0.71). Therefore, in this version, it makes sense to note an association with maternal BMI specifically.
The main weakness of the study comes from its negative results, which the authors emphasize as their primary conclusion: that previously reported associations of childhood BMI with anxiety, depression, and ADHD are not replicated using within-family MR methods. These claims do not seem justified by the evidence presented in this study. In fact, in every panel of figures 2 and 3, the error bars for the within-family MR analysis encompass the estimates for both the regression analysis and the traditional MR analysis, suggesting that the within-family analysis provides no evidence one way or another about which of these analyses is more accurate. More generally, in order to convincingly claim that there is no causal relationship between two traits, an MR study must argue that the study would be powered to detect a relationship if one existed. Within-family MR methods are known to have less power to detect associations and less precision to estimate effect sizes than traditional MR methods or traditional observational epidemiology methods, so it is not sufficient to show that these other methods have power to detect the association. To make this kind of claim, it is necessary to include some kind of power analysis, such as a simulation study or analytic power calculations, and likely also a positive control to show that this method does have power to detect known effects in this cohort.
We agree that it is imperative that negative (i.e. “non-significant”) results are correctly interpreted - it is just as important to discover what is unlikely to affect emotional and behavioural outcomes as what does affect them. Negative results (non-significant estimates) are neither a weakness nor strength of the study, but simply reflect the estimation error in our analysis of the data. The key question is whether our within-family MR estimates are sufficiently powered to detect effect sizes of interest or rule out clinically meaningful effect sizes – or are they simply too imprecise to draw any conclusions? As the reviewer suggests, one way to address this is via a post-hoc power calculation. We consider post-hoc power calculations redundant, since all the information about the power of our analysis is reflected in the standard errors and reported confidence intervals. Moreover, any post-hoc power calculation will be necessarily approximate compared to using the standard errors and confidence intervals which we report.
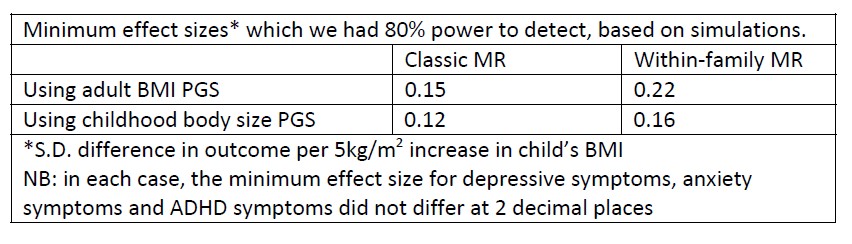
Despite these methodological reservations, we have conducted simulations to estimate the power of our within-family models (the R code is included at the end of this document). These simulations indicate that we do have sufficient power to detect the size of effects seen for depressive symptoms and ADHD in models using the adult BMI PGS. They also indicate that we cannot rule out smaller effects for non-significant associations (e.g., for the impact of the child’s BMI on anxiety). Naturally, this is entirely consistent with the width of the confidence intervals reported in results tables and in Figures 1 and 2. However, although power calculations are important when planning a study, they make little contribution to interpretation once a study has been conducted and confidence intervals are available (e.g., https://psyarxiv.com/tcqrn/). For this reason, we comment on these simulations in this response to reviewers but do not include them in the manuscript or supplementary materials. At the same time, we have changed the language used in the manuscript to be clearer that the results were imprecise and that values contained within the confidence limits cannot be ruled out.
For example, the discussion now includes the following:
‘However, within-family MR estimates using the childhood body size PGS are still consistent with small effects of the child’s BMI on all outcomes, with upper confidence limits around a 0.2 standard-deviation increase in the outcome per 5kg/m2 increase in BMI.’
And the conclusion of the paper now reads:
‘Our results suggest that genetic variation associated with BMI in adulthood affects a child’s depressive and ADHD symptoms, but genetic variation associated with recalled childhood body size does not substantially affect these outcomes. There was little evidence that BMI affects anxiety. However, our estimates were imprecise, and these differences may be due to estimation error. There was little evidence that parental BMI affects a child’s ADHD or anxiety symptoms, but factors associated with maternal BMI may independently influence a child’s depressive symptoms. Genetic studies using unrelated individuals, or polygenic scores for adult BMI, may have overestimated the causal effects of a child’s own BMI.’
Regarding a positive control: for analyses of BMI in adults, suitable positive controls would include directly measured biomarkers such as fat mass or blood pressure or reported medical outcomes like type 2 diabetes. In adolescents and younger adults, age at menarche or other measures of puberty can be used, as these are reliably influenced by BMI. However, the age of the participants for whom within-family effects are being estimated (8 years), together with the lack of any biomarkers such as fat mass (due to the questionnaire-based survey design) mean no suitable measures are available.
Reviewer #3 (Public Review):
Higher BMI in childhood is correlated with behavioral problems (e.g. depression and ADHD) and some studies have shown that this relationship may be causal using Mendelian Randomization (MR). However, traditional MR is susceptible to bias due to population stratification, assortative mating, and indirect effects (dynastic effects). To address this issue, Hughes et al. use within-family MR, which should be immune to the above-listed problems. They were unable to find a causal relationship between children's BMI and depression, anxiety, or ADHD. They do, however, report a causal effect of mother's BMI on depression in their children. They conclude that the causal effect of children's BMI on behavioral phenotypes such as depression and anxiety, if present, is very small, and may have been overestimated in previous studies. The analyses have been carried out carefully in a large sample and the paper is presented clearly. Overall, their assertions are justified but given that the conclusions mostly rest on an absence of an effect, I would like to see more discussion on statistical power.
- The authors show that the estimates of within-family MR are imprecise. It would be helpful to know how much power they have for estimating effect sizes reported previously given their sample size.
As discussed in response to a comment from reviewer 2, the power of our results is already indicated by our standard errors and confidence intervals. Nevertheless, we conducted simulations to estimate the size of effects which we had 80% power to detect. Results, presented below, are consistent with our main results. As discussed in response to a comment from reviewer 2, we consider post-hoc power calculations redundant when standard errors and confidence intervals are reported; for this reason, we include this information in the response to reviewers but not the manuscript itself.
- They used the correlation between PGS and BMI to support the assertion that the former is a strong instrument. Were the reported correlations calculated across all individuals? Since we know that stratification, assortative mating, and indirect effects can inflate these correlations, perhaps a more unbiased estimate would be the proportion of children's BMI variance explained by their PGS conditioned on the parents' PGS. This should also be the estimate used in power calculations.
The manuscript has been updated to quote Sanderson-Windmeijer conditional R2 values: the proportion of BMI variance explained by the BMI PGS for each member of a trio, conditional on the PGS of the other members of the trio, and all genetic covariates included in within-family models. Similarly, we now show Sanderson-Windmeijer conditional F-statistics for a model including the child, mother, and father’s BMI instrumented by the child, mother, and father’s PGS.
- In testing the association of mothers' and fathers' BMI with children's symptoms, the authors used a multivariable linear regression conditioning on the child's own BMI. Was the other parent's BMI (either by itself or using the polygenic score) included as a covariate in the multivariable and MR models? This was not entirely clear from the text or from Fig. 2. I suspect that if there were assortative mating on BMI in the parent's generation, the effect of any one parent's BMI on the child's symptoms might be inflated unless the other parent's BMI was included as a covariate (assuming both mother's and father's BMI affect the child's symptoms).
Non-genetic models include both the mother and father’s phenotypic BMI as well as the child’s, allowing estimation of conditional effects of all three. This controls for assortative mating as noted by the reviewer. This was not previously clear - all relevant text and figure captions have been updated to clarify this.
- They report no evidence of cross-trait assortative mating in the parents generation. The power to detect cross-trait assortative mating in the parents' generation using PGS would depend on the actual strength of assortative mating and the respective proportions of trait variance explained by PGS. Could the authors provide an estimate of the power for this test in their sample?
We have updated the discussion of assortative mating (in both the results and the discussion section) to note possible limitations of power and clarify that that this approach to examining assortment may not capture its full extent.
The relevant part of the results section now reads:
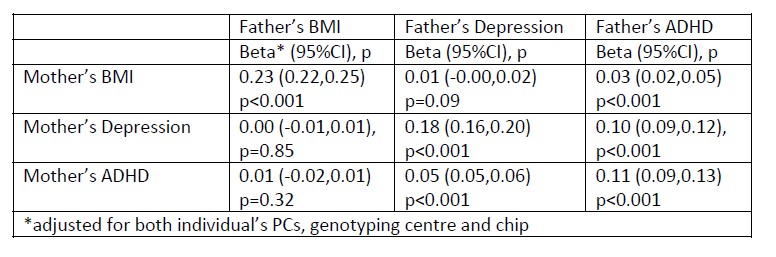
“In the parents’ generation, phenotypes were associated within parental pairs, consistent with assortative mating on these traits (Appendix 1 – Table 5). Adjusted for ancestry and other genetic covariates, maternal and paternal BMI were positively associated (beta: 0.23, 95%CI: 0.22,0.25, p<0.001), as were maternal and paternal depressive symptoms (beta: 0.18, 95%CI: 0.16,0.20, p<0.001), and maternal and paternal ADHD symptoms (beta: 0.11, 95%CI: 0.09,0.13, p<0.001). Consistent with cross-trait assortative mating, there was an association of mother’s BMI with father’s ADHD symptoms (beta: 0.03, 95%CI: 0.02,0.05, p<0.001) and mother’s ADHD symptoms with father’s depressive symptoms (beta: 0.05,95%CI: 0.05,0.06, p<0.001). Phenotypic associations can reflect the influence of one partner on another as well as selection into partnerships, but regression models of paternal polygenic scores on maternal polygenic scores also pointed to a degree of assortative mating. Adjusted for ancestry and genotyping covariates, there were small associations between parents’ BMI polygenic scores (beta: 0.01, 95%CI: 0.00,0.02, p=0.02 for the adult BMI PGS, and beta: 0.01, 95%CI: 0.00,0.02, p=0.008 for the childhood body size PGS), and of the mother’s childhood body size PGS with the father’s ADHD PGS (beta: 0.01, 95%CI: 0.00,0.02, p=0.03). We did not detect associations with pairs of other polygenic scores, which may be due to insufficient statistical power.”
And the relevant part of the discussion section now reads:
“We found some genomic evidence of assortative mating for BMI, and cross-trait assortative mating between BMI and ADHD, but not between other traits. However, associations between polygenic scores, which only capture some of the genetic variation associated with these phenotypes, may not capture the full extent of genetic assortment on these traits.”
- Are the actual phenotypes (BMI, depression or ADHD) correlated between the parents? If so, would this not suffice as evidence of cross-trait assortative mating? It is known that the genetic correlation between parents as a result of assortative mating is a function of the correlation in their phenotypes and the heritabilities underlying the two traits (e.g., see Yengo and Visscher 2018). An alternative way to estimate the genetic correlation between parents without using PGS (which is noisy and therefore underpowered) would be to use the phenotypic correlation and heritability estimated using GREML or LDSC. Perhaps this is outside the scope of the paper but I would like to hear the author's thoughts on this.
Associations between maternal and paternal phenotypes are consistent with a degree of assortative mating (shown below). These results have added to Appendix 1 - Table 5, which also shows associations between maternal and paternal polygenic scores, and methods and results updated accordingly (see quoted text in response to the comment above). For comparability, both sets of results are based on regression models adjusting for the mother’s and father’s ancestry PCs and genotyping covariates. We agree that analysis of assortative mating using GREML or LDSC is out of scope for this paper. As noted above, we have updated the discussion to acknowledge the limitations of the approach taken:
‘We found some genomic evidence of assortative mating for BMI, and cross-trait assortative mating between BMI and ADHD, but not between other traits. However, associations between polygenic scores, which only capture some of the genetic variation associated with these phenotypes, may not capture the full extent of genetic assortment on these traits.’
- It would be helpful to include power calculations for the MR-Egger intercept estimates.
As with our response to the comments above, post-hoc power calculations are redundant, as all the information about the power of our analysis, including the MR-Egger is indicated by the standard errors and confidence intervals. MR-Egger is less precise than other estimators, as is made clear from the wide confidence intervals reported in the relevant tables (Appendix 1 - Tables 8 and 9). However, we have now updated the discussion to give more weight to this as a limitation. The discussion of pleiotropy in the final paragraph of the discussion now reads:
‘While robustness checks found little evidence of pleiotropy, these methods rely on assumptions. Moreover, MR-Egger is known to give imprecise estimates (Burgess and Thompson 2017), and confidence intervals from MR-Egger models were wide. Thus, pleiotropy cannot be ruled out.’
Similarly, we have updated the relevant line of the results section, which now reads:
‘MR-Egger models found little evidence of horizontal pleiotropy, although MR-Egger estimates were imprecise (Appendix 1 - Tables 8 and 9).’
- Finally, what is the correlation between PGS and genetic PCs/geography in their sample? A correlation might provide evidence to support the point that classic MR effects are inflated due to stratification.
Figures presenting the association of the child’s BMI polygenic scores and their PCs have been added to the supplementary information as Appendix 1 - Figure 2 and Appendix 1 - Figure 3. Consistent with an influence of residual stratification, a regression of the child’s BMI polygenic scores against their ancestry PCs (adjusting for genotyping centre and chip) found that 7 of the 20 PCs were associated at p<0.05 with the adult BMI PGS, and 8 of 20 with the childhood body size PGS (under the null hypothesis, we would expect one association in each case). When parental polygenic scores were added to the models, these associations attenuated towards to null.
-

Evaluation Summary:
This paper uses a new statistical approach called within family Mendelian randomization and asserts that claims of childhood BMI affecting a range of psychiatric traits are unfounded and were mainly caused by confounders that this new approach is able to better identify and control for. They do find a role for maternal BMI on a child's risk for developing depression. The main issue raised is that they do not convincingly show if they do not replicate the old association of childhood BMI with a range of psychiatric traits due to their technique simply having lower power to detect the signal or due to a true lack of this effect.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #2 …
Evaluation Summary:
This paper uses a new statistical approach called within family Mendelian randomization and asserts that claims of childhood BMI affecting a range of psychiatric traits are unfounded and were mainly caused by confounders that this new approach is able to better identify and control for. They do find a role for maternal BMI on a child's risk for developing depression. The main issue raised is that they do not convincingly show if they do not replicate the old association of childhood BMI with a range of psychiatric traits due to their technique simply having lower power to detect the signal or due to a true lack of this effect.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #2 agreed to share their name with the authors.)
-

Reviewer #1 (Public Review):
The manuscript describes Mendelian Randomization (MR) analyses aimed at determining what, if any, causal effect body mass index (BMI) has on childhood emotional problems: depression, anxiety, and attention-deficit and hyperactivity disorder (ADHD) at age 8. To do this, the study leverages genetic association results on BMI to construct a genetic 'instrument', called a polygenic score, that predicts BMI. They use this score to see if the genetic predictor of BMI also predicts childhood emotional problems. What distinguishes this study from typical MR studies is that they use a large sample of 26,370 children with genotype data available for the child and both parents. This enables them to use within-family MR: within-family MR uses the parental genotypes as controls to remove confounding factors. Because …
Reviewer #1 (Public Review):
The manuscript describes Mendelian Randomization (MR) analyses aimed at determining what, if any, causal effect body mass index (BMI) has on childhood emotional problems: depression, anxiety, and attention-deficit and hyperactivity disorder (ADHD) at age 8. To do this, the study leverages genetic association results on BMI to construct a genetic 'instrument', called a polygenic score, that predicts BMI. They use this score to see if the genetic predictor of BMI also predicts childhood emotional problems. What distinguishes this study from typical MR studies is that they use a large sample of 26,370 children with genotype data available for the child and both parents. This enables them to use within-family MR: within-family MR uses the parental genotypes as controls to remove confounding factors. Because offspring genotype is randomly assigned given parental genotype, controlling for parental genotype removes bias due to gene-environment correlation and assortative mating.
The authors find that 'classic MR' (i.e. without controls for parental genotypes) gives evidence that higher BMI increases depressive symptoms and ADHD symptoms in children. However, when controlling for parental genotype (within-family MR), the estimates become smaller and are no longer statistically significant. While this is consistent with 'classic MR' being confounded due to gene-environment correlation and/or assortative mating, the within family MR analysis is less powerful (i.e,. considerable uncertainty about the effect remains) so it is hard to draw any strong conclusions about whether there is or is not an effect of BMI on childhood emotional problems.
This study provides further evidence that MR analyses that do not control for parental genotypes can be biased and conclusions drawn from these analyses should not be taken at face value. However, the fact that there is still a high degree of uncertainty in the within-family MR estimates despite having a large sample of children with genotyped parents implies that, for many hypotheses, much larger samples with genotyped parents will be needed to conduct well-powered within-family MR analyses. Further studies could also interrogate what aspects of the environment explain the observed correlation between parental genotype and offspring emotional problems.
-

Reviewer #2 (Public Review):
This study evaluates the causal relationship between childhood obesity on the one hand, and childhood emotional and behavioral problems on the other. It applies Mendelian Randomization (MR), a family of methods in statistical genetics that uses genetic markers to break the symmetry between correlated traits, allowing inference of causation rather than mere correlation. The authors argue convincingly that previous studies of these traits, both those using non-genetic observational epidemiology methods and those using standard MR methods, may be confounded by demographic effects and familial effects. One possible example of this kind of confounding is that the idea that obesity in parents may contribute to emotional and behavioral problems in children; another is the idea that adults with emotional and …
Reviewer #2 (Public Review):
This study evaluates the causal relationship between childhood obesity on the one hand, and childhood emotional and behavioral problems on the other. It applies Mendelian Randomization (MR), a family of methods in statistical genetics that uses genetic markers to break the symmetry between correlated traits, allowing inference of causation rather than mere correlation. The authors argue convincingly that previous studies of these traits, both those using non-genetic observational epidemiology methods and those using standard MR methods, may be confounded by demographic effects and familial effects. One possible example of this kind of confounding is that the idea that obesity in parents may contribute to emotional and behavioral problems in children; another is the idea that adults with emotional and behavioral issues may be more likely to have children with partners who are obese, and vice-versa. They then make use of a recently proposed "within-family" MR method, which should effectively control for these confounders, at the cost of higher uncertainty in the estimated effect size, and therefore lower power to detect small effects. They report that none of the previously reported associations of childhood BMI with anxiety, depression, or ADHD are replicated using the within-family MR method, and that in the case of depression the primary association appears to be with maternal BMI rather than the child's own BMI.
This argument that these confounders may affect these phenotypes is fairly sound, and within-family MR should indeed do a good job of controlling for them. I do not see any major issues with the cohort itself or the choice of genetic instruments. I also do not see any major issues with the definitions or ascertainment of the phenotypes studied, though I am not an expert on any of these phenotypes in particular. I am especially satisfied with the series of analyses demonstrating that the results are robust to many variations of MR methodology. Overall, I think the positive result this study reports is very credible: that the known association between childhood BMI and depression is likely primarily due to an effect of maternal BMI rather than the child's own BMI (though given that paternal BMI has a similar effect size with only a slightly wider confidence interval, I would instead say that the effect is from parental BMI generally, not specifically maternal.)
The main weakness of the study comes from its negative results, which the authors emphasize as their primary conclusion: that previously reported associations of childhood BMI with anxiety, depression, and ADHD are not replicated using within-family MR methods. These claims do not seem justified by the evidence presented in this study. In fact, in every panel of figures 2 and 3, the error bars for the within-family MR analysis encompass the estimates for both the regression analysis and the traditional MR analysis, suggesting that the within-family analysis provides no evidence one way or another about which of these analyses is more accurate. More generally, in order to convincingly claim that there is no causal relationship between two traits, an MR study must argue that the study would be powered to detect a relationship if one existed. Within-family MR methods are known to have less power to detect associations and less precision to estimate effect sizes than traditional MR methods or traditional observational epidemiology methods, so it is not sufficient to show that these other methods have power to detect the association. To make this kind of claim, it is necessary to include some kind of power analysis, such as a simulation study or analytic power calculations, and likely also a positive control to show that this method does have power to detect known effects in this cohort.
-

Reviewer #3 (Public Review):
Higher BMI in childhood is correlated with behavioral problems (e.g. depression and ADHD) and some studies have shown that this relationship may be causal using Mendelian Randomization (MR). However, traditional MR is susceptible to bias due to population stratification, assortative mating, and indirect effects (dynastic effects). To address this issue, Hughes et al. use within-family MR, which should be immune to the above-listed problems. They were unable to find a causal relationship between children's BMI and depression, anxiety, or ADHD. They do, however, report a causal effect of mother's BMI on depression in their children. They conclude that the causal effect of children's BMI on behavioral phenotypes such as depression and anxiety, if present, is very small, and may have been overestimated in …
Reviewer #3 (Public Review):
Higher BMI in childhood is correlated with behavioral problems (e.g. depression and ADHD) and some studies have shown that this relationship may be causal using Mendelian Randomization (MR). However, traditional MR is susceptible to bias due to population stratification, assortative mating, and indirect effects (dynastic effects). To address this issue, Hughes et al. use within-family MR, which should be immune to the above-listed problems. They were unable to find a causal relationship between children's BMI and depression, anxiety, or ADHD. They do, however, report a causal effect of mother's BMI on depression in their children. They conclude that the causal effect of children's BMI on behavioral phenotypes such as depression and anxiety, if present, is very small, and may have been overestimated in previous studies. The analyses have been carried out carefully in a large sample and the paper is presented clearly. Overall, their assertions are justified but given that the conclusions mostly rest on an absence of an effect, I would like to see more discussion on statistical power.
The authors show that the estimates of within-family MR are imprecise. It would be helpful to know how much power they have for estimating effect sizes reported previously given their sample size.
They used the correlation between PGS and BMI to support the assertion that the former is a strong instrument. Were the reported correlations calculated across all individuals? Since we know that stratification, assortative mating, and indirect effects can inflate these correlations, perhaps a more unbiased estimate would be the proportion of childrens' BMI variance explained by their PGS conditioned on the parents' PGS. This should also be the estimate used in power calculations.
In testing the association of mothers' and fathers' BMI with children's symptoms, the authors used a multivariable linear regression conditioning on the child's own BMI. Was the other parent's BMI (either by itself or using the polygenic score) included as a covariate in the multivariable and MR models? This was not entirely clear from the text or from Fig. 2. I suspect that if there were assortative mating on BMI in the parent's generation, the effect of any one parent's BMI on the child's symptoms might be inflated unless the other parent's BMI was included as a covariate (assuming both mother's and father's BMI affect the child's symptoms).
They report no evidence of cross-trait assortative mating in the parents generation. The power to detect cross-trait assortative mating in the parents' generation using PGS would depend on the actual strength of assortative mating and the respective proportions of trait variance explained by PGS. Could the authors provide an estimate of the power for this test in their sample?
Are the actual phenotypes (BMI, depression or ADHD) correlated between the parents? If so, would this not suffice as evidence of cross-trait assortative mating? It is known that the genetic correlation between parents as a result of assortative mating is a function of the correlation in their phenotypes and the heritabilities underlying the two traits (e.g., see Yengo and Visscher 2018). An alternative way to estimate the genetic correlation between parents without using PGS (which is noisy and therefore underpowered) would be to use the phenotypic correlation and heritability estimated using GREML or LDSC. Perhaps this is outside the scope of the paper but I would like to hear the author's thoughts on this.
It would be helpful to include power calculations for the MR-Egger intercept estimates.
Finally, what is the correlation between PGS and genetic PCs/geography in their sample? A correlation might provide evidence to support the point that classic MR effects are inflated due to stratification.
-