Mapping brain-wide excitatory projectome of primate prefrontal cortex at submicron resolution and comparison with diffusion tractography
Curation statements for this article:-
Curated by eLife
Evaluation Summary:
This paper will be of broad interest to readers who study anatomical connections of the brain. It demonstrates the efficacy of a cutting-edge viral tracing technique in mapping excitatory projections in macaque monkeys. The work describes the generation of a projectome from the macaque vlPFC cortex across the rest of the brain using AAV2/9-CaMKIIa-Tau-GFP labeling and imaging with high-throughput serial two-photon tomography. The comparison with imaging techniques available in humans (diffusion tractography) will also be of interest to research in human brain anatomy.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #1 agreed to share their name with the authors.)
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Resolving trajectories of axonal pathways in the primate prefrontal cortex remains crucial to gain insights into higher-order processes of cognition and emotion, which requires a comprehensive map of axonal projections linking demarcated subdivisions of prefrontal cortex and the rest of brain. Here, we report a mesoscale excitatory projectome issued from the ventrolateral prefrontal cortex (vlPFC) to the entire macaque brain by using viral-based genetic axonal tracing in tandem with high-throughput serial two-photon tomography, which demonstrated prominent monosynaptic projections to other prefrontal areas, temporal, limbic, and subcortical areas, relatively weak projections to parietal and insular regions but no projections directly to the occipital lobe. In a common 3D space, we quantitatively validated an atlas of diffusion tractography-derived vlPFC connections with correlative green fluorescent protein-labeled axonal tracing, and observed generally good agreement except a major difference in the posterior projections of inferior fronto-occipital fasciculus. These findings raise an intriguing question as to how neural information passes along long-range association fiber bundles in macaque brains, and call for the caution of using diffusion tractography to map the wiring diagram of brain circuits.
Article activity feed
-

-

Author Response:
Reviewer #1 (Public Review):
The authors report the generation of a mesoscale excitatory projectome from the ventrolateral prefrontal cortex (vlPFC) in the macaque brain by using AAV2/9-CaMKIIa-Tau-GFP labeling and imaging with high-throughput serial two-photon tomography. They present a novel data pipeline that integrates the STP data with macroscopic dMRI data from the same brain in a common 3D space, achieving a direct comparison of the two tracing methods. The analysis of the data revealed an interesting discrepancy between the high resolution STP data and the lower resolution dMRI data with respect to the extent of the frontal lobe projection through the inferior fronto-occipital fasciculus (IFOF) - the longest associative axon bundle in the human brain.
The authors report the generation of a mesoscale …
Author Response:
Reviewer #1 (Public Review):
The authors report the generation of a mesoscale excitatory projectome from the ventrolateral prefrontal cortex (vlPFC) in the macaque brain by using AAV2/9-CaMKIIa-Tau-GFP labeling and imaging with high-throughput serial two-photon tomography. They present a novel data pipeline that integrates the STP data with macroscopic dMRI data from the same brain in a common 3D space, achieving a direct comparison of the two tracing methods. The analysis of the data revealed an interesting discrepancy between the high resolution STP data and the lower resolution dMRI data with respect to the extent of the frontal lobe projection through the inferior fronto-occipital fasciculus (IFOF) - the longest associative axon bundle in the human brain.
The authors report the generation of a mesoscale excitatory projectome from the ventrolateral prefrontal cortex (vlPFC) in the macaque brain by using AAV2/9-CaMKIIa-Tau-GFP labeling and imaging with high-throughput serial two-photon tomography. They also present a novel data pipeline that integrates the STP data with macroscopic dMRI data from the same brain in a common 3D space, achieving a direct comparison of the two tracing methods. Overall the paper can serve as a how to example for analyzing large non-human primate brain data, though some parts of the paper can be improved and the interpretation of the data should also be further strengthened.
We thank the reviewer for his positive evaluation of our manuscript.
The methodological part should include more detail on image acquisition - speed of imaging, pixel residence time, total time for data acquisition of a single brain and data sizes. Also the time and hardware needed for the computational analysis should be included, including the registration to the common reference and the running time for the machine learning predictions - this should also include the F score for the axon detection.
We thank the reviewer for pointing out these vital issues. We have added these technical details in the resubmitted manuscript.
“High x-y resolution (0.95 μm/pixel) serial 2D images were acquired in the coronal plane at a z-interval of 200 μm across the entire macaque brain. The scanning time of a single field-of-view which contains 1024 by 1024 pixels was 1.629 s (i.e., pixel residence time was ~1.6 μs), as resulted in a continuous ~1 month scanning and ~5 TB STP tomography data for a single monkey brain.”
“The data analysis was undertaken on a compute cluster with a 3.1 - 3.3 GHz 248 core CPU, 2.8 T of RAM, and 17472 CUDA cores.”
“The total computational time for the machine learning predictions in one macaque brain was ~ 1.5 months.”
“To evaluate overall classifier performance, the precision–recall F measure, also called F-score, was computed by using additional four labeled images as test sets. Higher accuracy performance achieved by the classifier often yield higher F-scores (94.41% ± 1.99%, mean ± S.E.M.).”
“For registration to the 3D common space, it took half an hour approximately.”
The discrepancy between the high resolution STP data and the lower resolution dMRI data with respect to the extent of the frontal lobe projection through the inferior fronto-occipital fasciculus seems puzzling. One would expect that the STP data would reveal more detail not less.. One possibility is that the Tau-GFP does not diffuse throughout the full axon arborization of the PFC neurons, resulting in a technical artifact. Can this be excluded to support the functional significance of the current data?
We thank the reviewer for raising this important issue. We apologize for not providing sufficient details of the IFOF debate due to limited space and causing confusion. We have added literature background of the IFOF debate to the section of Introduction (also recommended by Reviewer #2). Thanks to the comments by Reviewer #2, the present finding provides direct support for the speculation that the IFOF of macaque monkeys may not exist in a mono-synaptic way.
The AAV construct encoding cytoskeletal GFP (Tau-GFP) was used here to label all processes of the infected neuron, including axons and synaptic terminals. About 3 weeks of post-surgery survival time are usually sufficient to label intracerebral circuits in rodents (Lanciego and Wouterlood, 2020). We have extended the survival time to 2-3 months in order to achieve adequate labeling of axonal fibers and terminals in macaques.
Regarding the extent of Tau-GFP diffuse, the STP images and high-resolution confocal microscopic analysis further showed differences in the morphology of axon fibers that populate the route and terminals of these axon fibers. Consistent with previous reports (Fuentes-Santamaria et al., 2009; Watakabe and Hirokawa, 2018), the axon fibers were thin and formed bouton-like varicosities in the terminal regions (MD, Figure 2—figure supplement 7D; caudate, Figure 2—figure supplement 7J; PFC, Figure 1—figure supplement 5A-D). Those results indicate that the Tau-GFP has reached axonal terminals.
References:
Fuentes-Santamaria V, Alvarado JC, McHaffie JG, Stein BE (2009) Axon Morphologies and Convergence Patterns of Projections from Different Sensory-Specific Cortices of the Anterior Ectosylvian Sulcus onto Multisensory Neurons in the Cat Superior Colliculus. Cereb Cortex 19:2902-2915.
Lanciego JL, Wouterlood FG (2020) Neuroanatomical tract-tracing techniques that did go viral. Brain Struct Funct 225:1193-1224.
Watakabe A, Hirokawa J (2018) Cortical networks of the mouse brain elaborate within the gray matter. Brain Struct Funct 223:3633-3652.
Reviewer #2 (Public Review):
The authors utilized viral vectors as neural tracers to delineate the connectivity map of the macaque vlPFC at the axonal level. There are three main goals of this study: 1) determine an effective viral vector for tract-tracing in the macaque brain, 2) delineate the detailed map of excitatory vlPFC projections to the rest of the brain, and 3) compare vlPFC connectivity between tracing and tractography results.
We thank the reviewer for his/her constructive comments, to which we respond below.
Accordingly, my comments are organized around each aim:
- This study demonstrates the advantage of viral tracing technique in targeting neuron type-specific pathways. The authors conducted injection experiments with three types of viral vectors and found success of AAV in labeling long-distance connections without causing fatal neurotoxicity in the monkey. This success extends the application of AAV from rodents to nonhuman primates. The fact that AAV specifically targets glutamatergic neurons makes it advantageous for mapping excitatory projections.
Although the labeling efficacy of each viral vector type is described in the text, Fig. 2 does not present a clear comparison across viral vectors, despite such comparison for a thalamic injection in Fig. 2S. Without a comparable graph to Fig. 2E, it is unclear to what extent the VSV and lentivirus failed in labeling long-distance pathways.
We thank the reviewer for the helpful suggestion. As suggested, we have added three new figures as Supplementary materials in the revised manuscript.
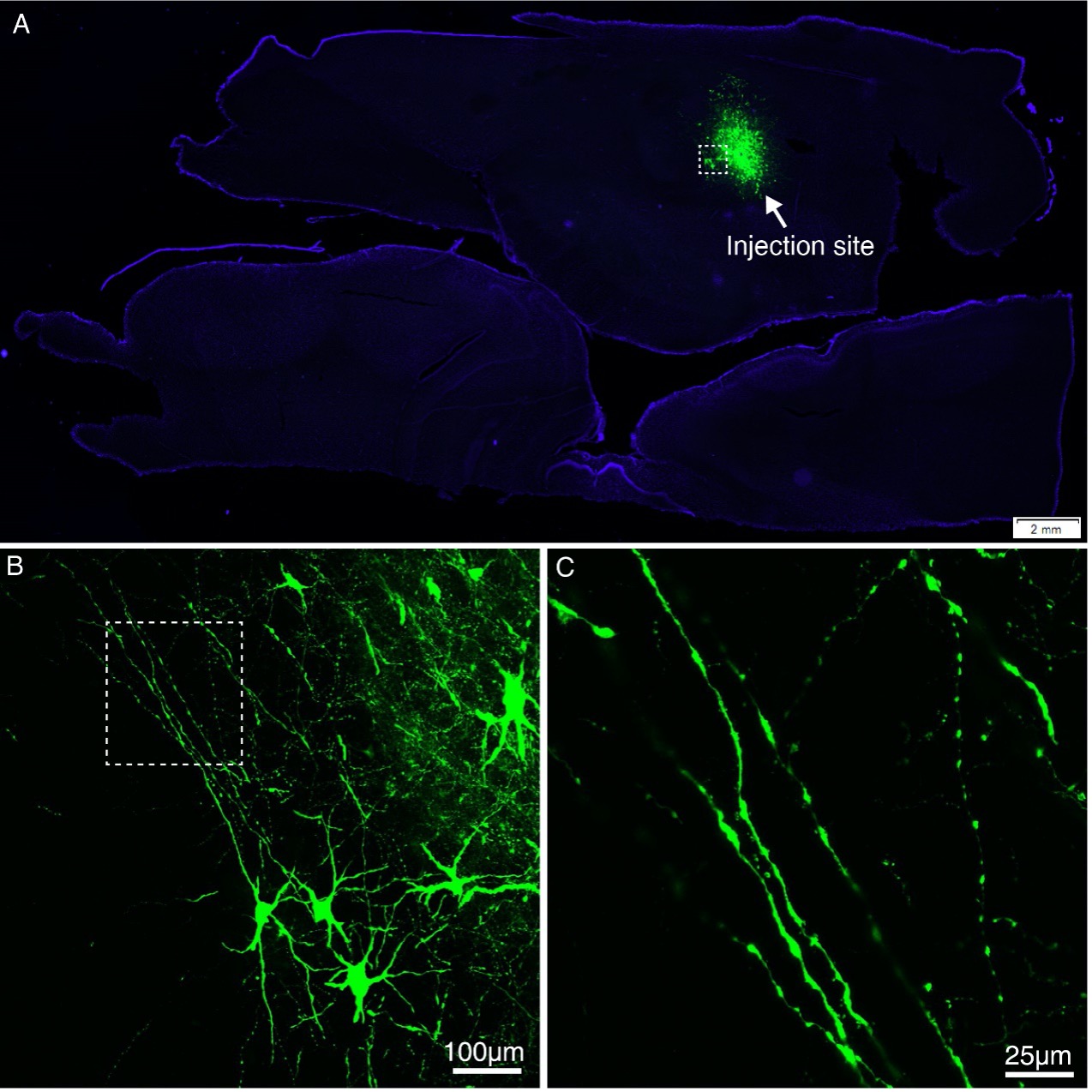
Figure 2—figure supplement 2. Expression of GFP using VSV-△G injected into MD thalamus of the macaque brain. (A) GFP-labeled neurons were found in the MD thalamus ~5 days after injection of VSV-△G encoding Tau-GFP. (B) A magnified view illustrating the morphology of GFP-labeled neurons in the area outlined with a white box in (A). (C) Higher magnification view of GFP-positive axons.
Figure 2—figure supplement 3. Expression of GFP using lentivirus injected into MD thalamus of the macaque brain. (A) Lentivirus construct was injected into the macaque thalamus and examined for transgene expression after ~9 months. (B) High power views of the dotted rectangle in panel A. (C) Magnified view of panel B. Note the presence of GFP-positive cells.
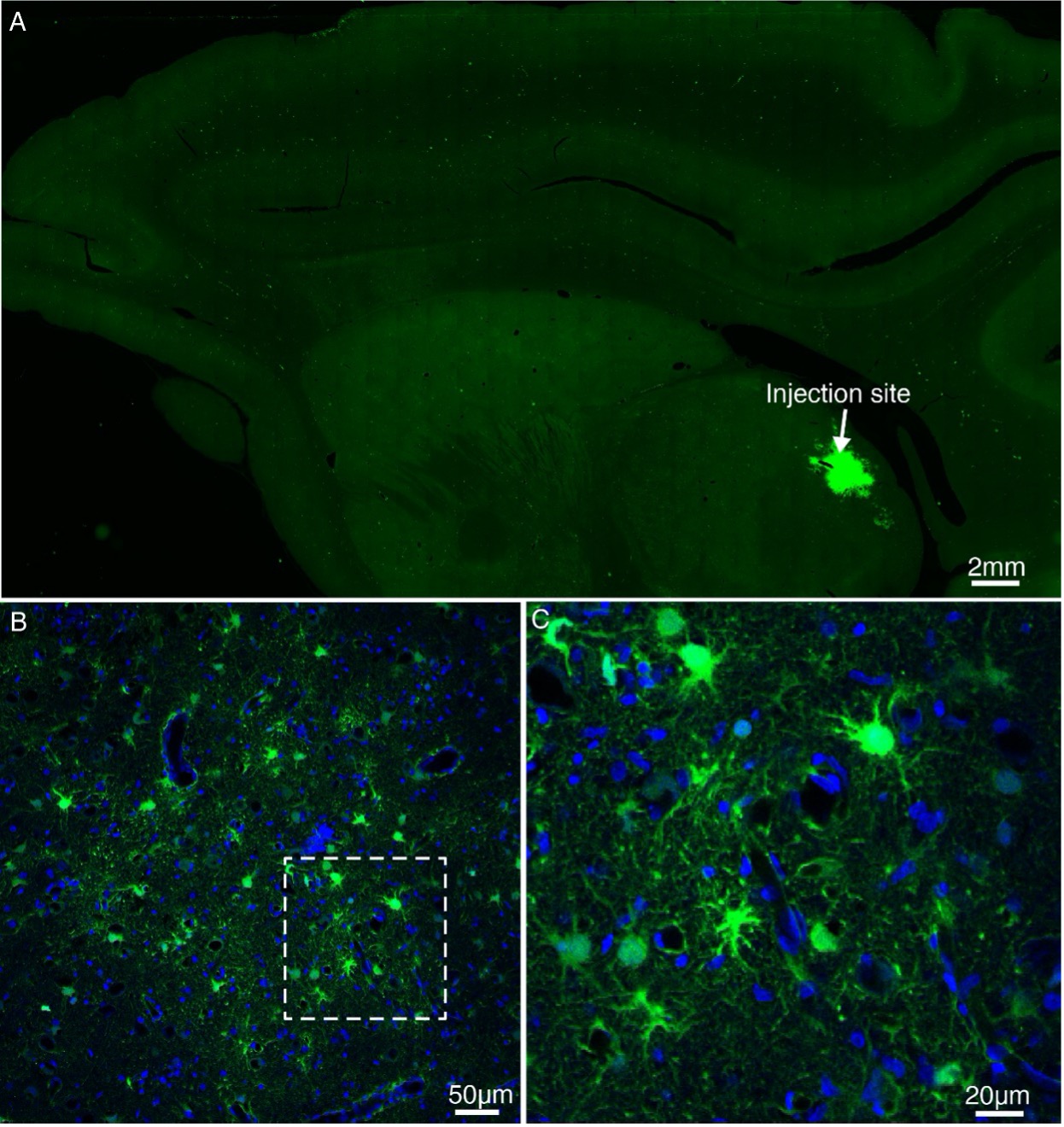
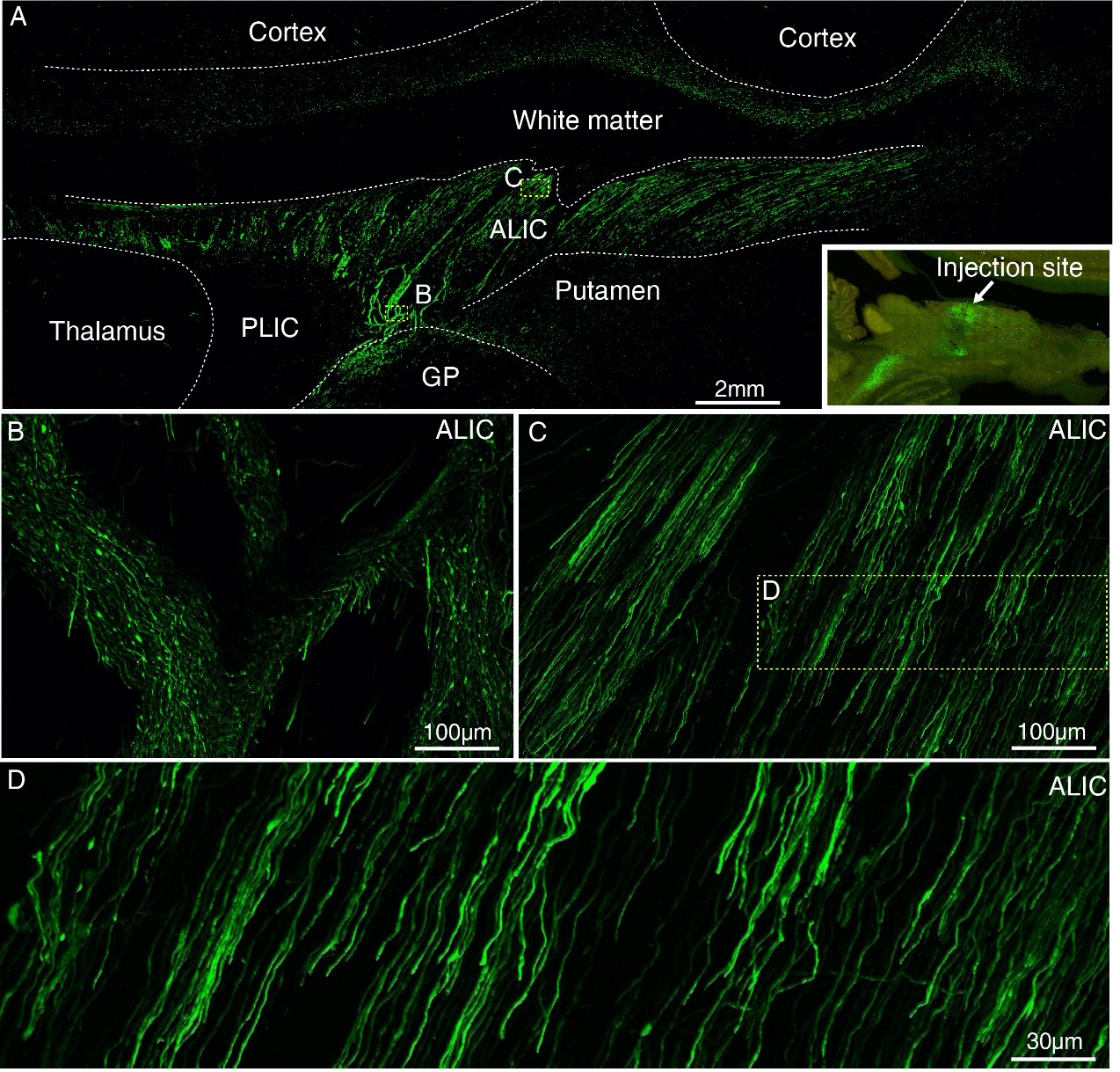
Figure 2—figure supplement 4. Expression of GFP using AAV2/9 injected into MD thalamus of the macaque brain. (A) GFP-labeled axons were observed in the subcortical regions ~42 days after injection of AAV2/9 encoding Tau-GFP in MD thalamus. The inset shows the injection site in MD thalamus. Two dashed line boxes enclose the regions of interest: frontal white matter and ALIC, whose GFP signal are magnified in (B) and (C), respectively. (D) Higher magnification view of GFP-positive axons.
- The authors quantified connectivity strength by the GFP signal intensity using a machine-learning algorithm. Both the quantitative approach and the resulting excitatory projection map are important contributions to advancing our knowledge of vlPFC connectivity.
However, several issues with the analysis lead to concerns about the connectivity result. First, the strength measure is based on axonal patterns in the terminal fields (which the authors refer to as "axon clusters"), detected by a machine-learning algorithm (page 25, lines 11-13). However, the actual synaptic connections are the small dot-looking signals in the background. These "green dots" are boutons on the dendritic trees. The density of boutons rather than the passing fibers reflects the density of synapses. The brief method description does not mention how the boutons are quantified, and it is unclear whether the signal was treated as the background noise and filtered out. Second, it is difficult for the reader to assess the robustness of the vlPFC connectivity patterns, due to these issues: i) It is unclear how many injection cases were used to generate the result reported in the subsection "Brain-wide excitatory projectome of vlPFC in macaques". The text mentions a singular "injection site" (page 8, line 12) and Fig. 4 shows a single site. However, there are three cases listed in Table 1. Is the result an average of all three cases? ii) Relatedly, it is unclear in which anatomical area the injection was placed for each case. Table 1 lists the site as "vlPFC" for all three cases, while the vlPFC contains areas 44, 45 and 12l. These areas have different projection patterns documented in the tract tracing literature. If different areas were injected in the three cases, they should be reported separately. iii) It is hard to compare the projection patterns with those reported in the literature. Conventionally, tract tracing studies report terminal fields by showing original labeling patterns in both cortical and subcortical regions without averaging within divided areas (see e.g. Petrides & Pandya, 2007, J Neurosci). It is hard to compare Fig. 3 with previous tract tracing studies to assess its robustness.
We thank the reviewer for his/her constructive comments, to which we respond below.
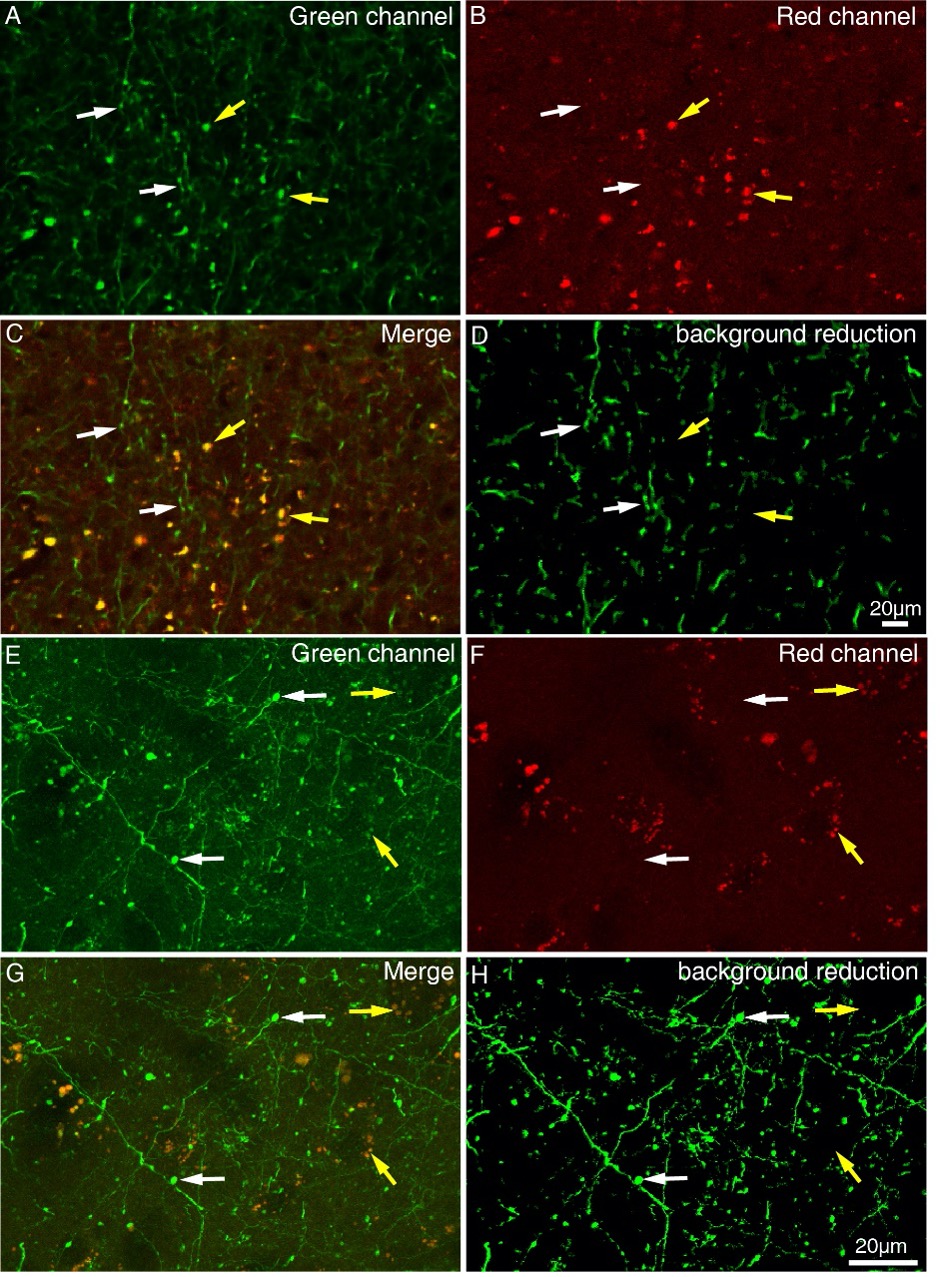
1). We appreciate the reviewer’s comment and sincerely apologize for not explaining this point clearly in our previous submission. The major concern is whether the axonal varicosities were likely to be treated as the background noise and removed by mistake. In fact, the dot-looking autofluorescence rather than the axonal varicosities were reduced through a machine-learning algorithm in segmentation. Hence we have provided new results and updated the “Materials and Methods” and “Discussion” sections in the revision accordingly.
“Fluorescent images of primate (Abe et al., 2017) brain often contain high-intensity dot-looking background signal caused by accumulation of lipofuscin. Thanks to the broad emission spectrum of lipofuscin, dot-looking background and GFP-positive axonal varicosities are easily distinguishable from each other. For instance (Figure 1—figure supplement 4), axonal varicosities can be selectively excited in green channel, while dot-looking background lipofuscin usually present in both green channel and red channel. During quantitative analysis, a machine learning algorithm was adopted to reliably segment the GFP labelled axonal fibers including axonal varicosities, and remove the lipofuscin background (Arganda-Carreras et al., 2017; Gehrlach et al., 2020).”
“One recent study compared results of terminal labelling using Synaptophysin-EGFP-expressing AAV (specifically labelling synaptic endings) with the cytoplasmic EGFP AAV (labelling axon fibers and synaptic endings). There was high correspondence between synaptic EGFP and cytoplasmic EGFP signals in target regions (Oh et al., 2014). Thus, we relied on quantifying GFP-positive pixels (containing signals from both axonal fibers and terminals) rather than the number of synaptic terminals, similarly done in recent reports (Oh et al., 2014; Gehrlach et al., 2020).”
Figure 1—figure supplement 4. Difference between axonal varicosities and dot-looking background. STP images (A-D) and high-resolution confocal images (E-H) were acquired in green channel and the red channel. Synaptic terminals (indicated by white arrows) can be specifically excited in green channel, while dot-looking background lipofuscin (indicated by yellow arrows) can be visualized both in green channel and red channel. (C and G) No colocalization was found between axonal varicosities and dot-looking background. Axonal varicosities were easily distinguished from dot-looking background in the merged image. (D and H) The dot-looking autofluorescence rather than the axonal varicosities was reduced through a machine-learning algorithm.
References:
Abe H, Tani T, Mashiko H, Kitamura N, Miyakawa N, Mimura K, Sakai K, Suzuki W, Kurotani T, Mizukami H, Watakabe A, Yamamori T, Ichinohe N (2017) 3D reconstruction of brain section images for creating axonal projection maps in marmosets. J Neurosci Methods 286:102-113.
Arganda-Carreras I, Kaynig V, Rueden C, Eliceiri KW, Schindelin J, Cardona A, Sebastian Seung H (2017) Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics 33:2424-2426.
Gehrlach DA, Weiand C, Gaitanos TN, Cho E, Klein AS, Hennrich AA, Conzelmann KK, Gogolla N (2020) A whole-brain connectivity map of mouse insular cortex. Elife 9.
Oh SW et al. (2014) A mesoscale connectome of the mouse brain. Nature 508:207-214.
2.1) We apologize for causing these confusions due to insufficient description in the main text. Now we have revised the description of the “Materials and Methods” section accordingly. Furthermore, we have made both the whole-brain serial two-photon data and high-resolution diffusion MRI data freely available to the community, as allows researchers in the field to perform further analyses that we have not done in the current study.
“Three samples were injected with AAV in vlPFC, and two of them were able to be imaged with STP. Unfortunately, one sample became “loose” and fell off from the agar block after several weeks of imaging. So, the quantitative results were not shown in Figure 3.”
2.2) We apologize for insufficient description of the precise location of the injection sites. We have revised the description of “Materials and Methods” section and provided a new figure to clarify the exact location of the injection sites.
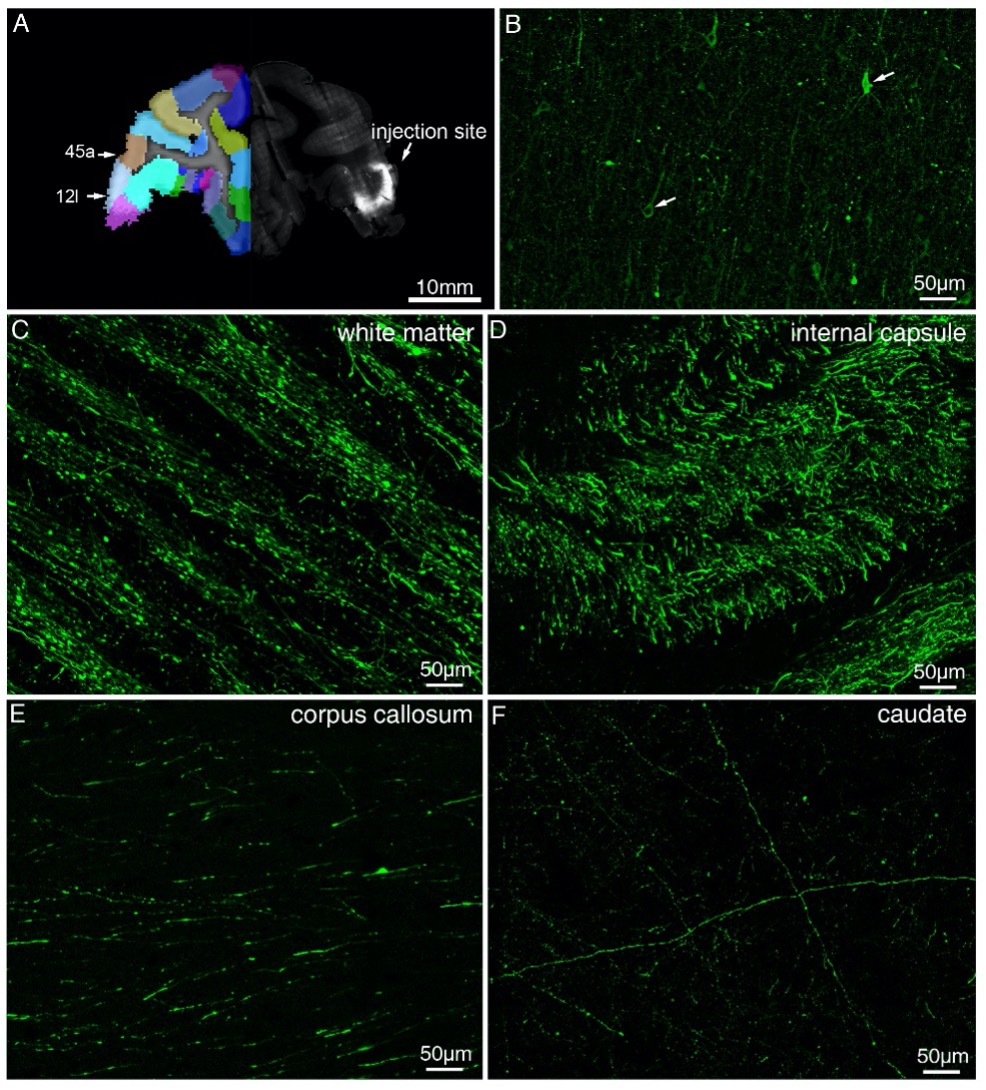
“Figure 3-4 and Figure 4—figure supplement 2-4 were derived from sample #8 with infected area in 45, 12l and 44 of vlPFC. Figure 1—figure supplement 6 was derived from sample #7 with infected area in 12l and 45 of vlPFC.”
Figure 1—figure supplement 6. Representative fluorescent images showing injection site and major tracts of sample #7. (A) STP image of the injection site in vlPFC are shown overlaid with the monkey brain template (left hand side), mainly spanning areas 12l and 45a. (B) Confocal image of the AAV infected neurons (indicated by white arrows). (C-F) Representative confocal images of major tracts originating from vlPFC.
2.3) We agree with the reviewer that most tract tracing studies report terminal fields by showing original labeling patterns. Several recent studies report the total volume of segmented GFP-positive pixels (Oh et al., 2014) or percentage of total labeled axons (Do et al., 2016; Gehrlach et al., 2020) to represent the connectivity strength, and other studies provide the projection density as well (Hunnicutt et al., 2016). We have provided both percentage of total labeled axons (Figure 3C right panel), projection density (Figure 3C left panel) and representative original fluorescent images (Figure. 4, Figure 4—figure supplement 2 and Figure 4—figure supplement 4) to demonstrate our projection data at different dimensions.
References:
Do JP, Xu M, Lee SH, Chang WC, Zhang S, Chung S, Yung TJ, Fan JL, Miyamichi K, Luo L, Dan Y (2016) Cell type-specific long-range connections of basal forebrain circuit. Elife 5.
Gehrlach DA, Weiand C, Gaitanos TN, Cho E, Klein AS, Hennrich AA, Conzelmann KK, Gogolla N (2020) A whole-brain connectivity map of mouse insular cortex. Elife 9.
Hunnicutt BJ, Jongbloets BC, Birdsong WT, Gertz KJ, Zhong H, Mao T (2016) A comprehensive excitatory input map of the striatum reveals novel functional organization. Elife 5.
Oh SW et al. (2014) A mesoscale connectome of the mouse brain. Nature 508:207-214.
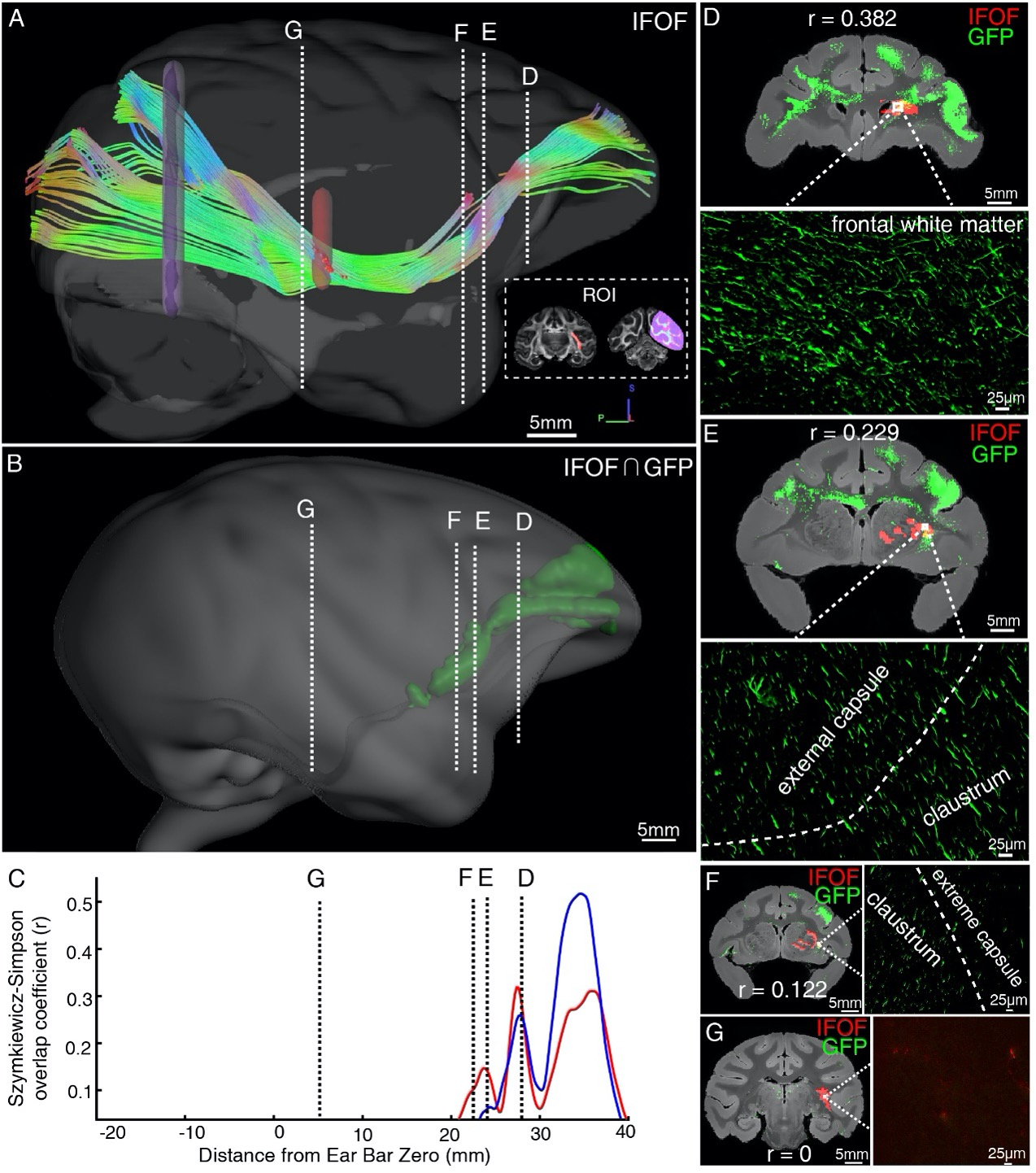
- Using the ground-truth from tract tracing to validate tractography results is a timely problem and this study showed promising consistency and discrepancy between the two modalities. Especially, the discrepancy between tracing and tractography data on the IFOF termination brings critical insights into a potential cross-species difference. The finding that IFOF does not reach the occipital cortex provides important support for the speculation that IFOF may not exist in monkeys (for a context of the IFOF debate see Schmahmann & Pandya, 2006, pp 445-446).
I have minor concerns regarding the statistical robustness of the tracing-tractography comparison. The authors compared the vlPFC-CC-contralateral tract instead of a global connectivity pattern without justification. Why omitting other major tracts that connect with vlPFC? In addition, the results are shown for only one monkey, while two monkeys went through both tracer injection and dMRI scans. It is unclear how the results were chosen or whether the data were averaged.
We apologize for not describing it clearly. The STP images were acquired in the coronal plane with high x-y resolution (0.95 μm/pixel), while the z resolution was relatively low (200 μm). The axonal connection information along z axis may be lost due to the present step size (relatively large) such that it is technically demanding to reconstruct the axonal density maps in sagittal or horizontal plane. Therefore, we focused on the vlPFC-CC-contralateral tract traveling along the coronal plane when quantifying the similarity coefficients along the anterior-posterior axis of the whole macaque brain, and omitted the tracts that were shown as dots in the coronal plane. We have revised it in the resubmitted manuscript.
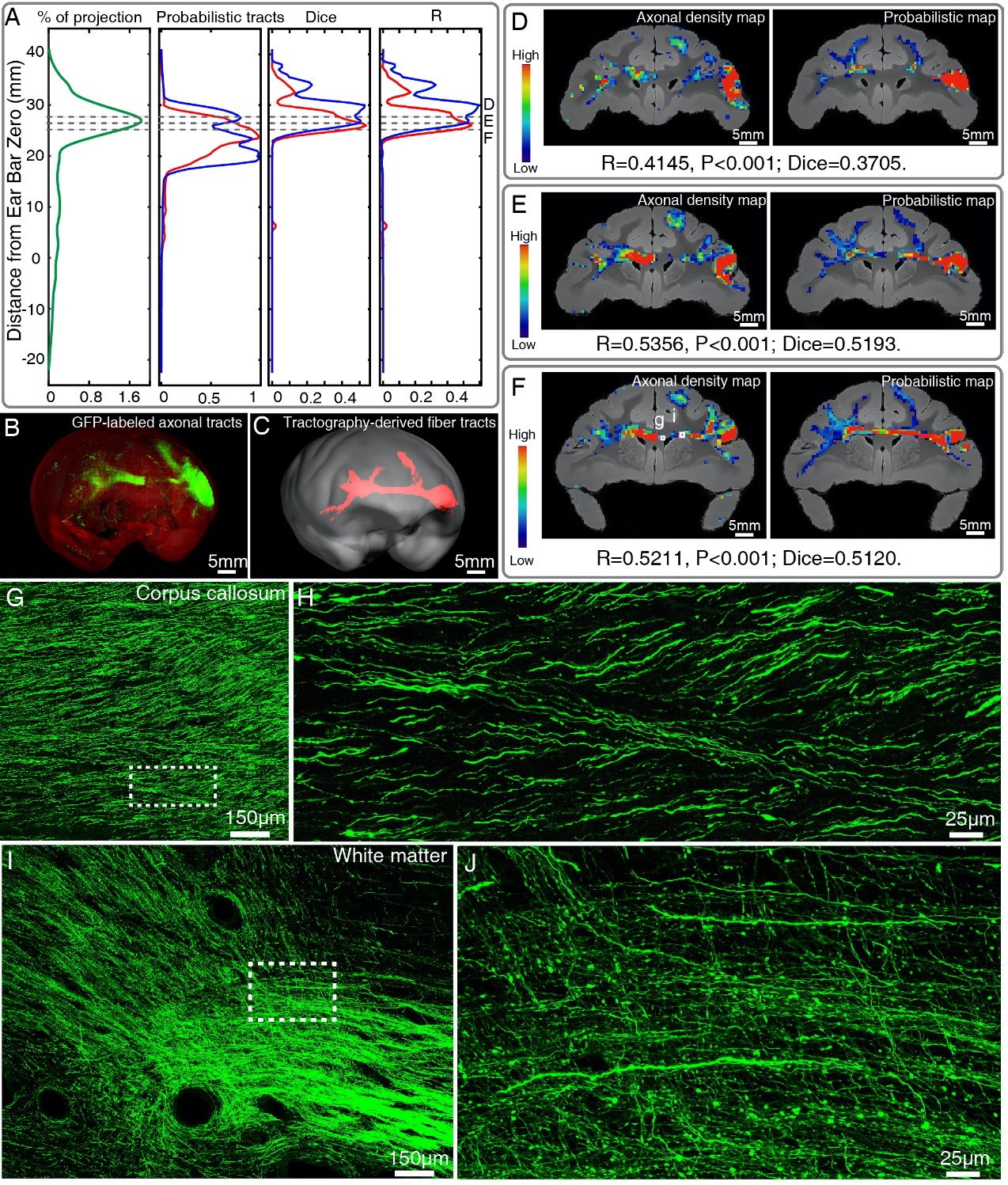
“GFP projection and probabilistic tract were plotted with the Dice coefficients and Pearson coefficients (R) along the anterior-posterior axis of the whole macaque brain. The Dice coefficients and Pearson coefficients were higher in dense projection regions, especially for the vlPFC-CC-contralateral tract (Figure 6A). To carry out a proof-of-principle investigation, we focused on the vlPFC-CC-contralateral tract that was reconstructed in 3D space by using STP and dMRI data, respectively.”
With regard to the demonstration of dMRI data, we apologize for not making it clear in previous version. We have already revised Figure 6 and Figure 7 so that dMRI scans from different macaque monkeys were shown separately.
Figure 6. Comparison of vlPFC connectivity profiles by STP tomography and diffusion tractography. (A) Percentage of projection, Probabilistic tracts, Dice coefficients and Pearson coefficients (R) were plotted along the anterior-posterior axis in the macaque brain. Blue and red colors indicate results of two dMRI data sets acquired from different macaque monkeys. (B, C) 3D visualization of the fiber tracts issued from the injection site in vlPFC to corpus callosum to the contralateral vlPFC by STP tomography and diffusion tractography. (D-F) Representative coronal slices of the diffusion tractography map and the axonal density map along the vlPFC-CC-contralateral tract, overlaid with the corresponding anatomical MR images. (G-J) GFP-labeled axon images as marked in Figure 6F were shown with magnified views. (H, J) correspond to high magnification images of the white boxes indicated in G and I, both of which presented a great deal of details about axonal morphology.
Figure 7. Illustration of the inferior fronto-occipital fasciculus by diffusion tractography and STP. (A) The fiber tractography of IFOF (lateral view). Two inclusion ROIs at the external capsule (pink) and the anterior border of the occipital lobe (purple) were used and shown on the coronal plane. The IFOF stems from the frontal lobe, travels along the lateral border of the caudate nucleus and external/extreme capsule, forms a bowtie-like pattern and anchors into the occipital lobe. (B) The reconstructed traveling course of IFOF based on vlPFC projectome was shown in 3D space. (C) The Szymkiewicz-Simpson overlap coefficients between 2D coronal brain slices of the dMRI-derived IFOF tract and vlPFC projections were plotted along the anterior-posterior axis of the macaque brain. Blue and red colors indicate results of two dMRI data sets acquired from different macaque monkeys. Four cross-sectional slices (D-G) along the IFOF tracts were arbitrarily chosen to demonstrate the spatial correspondence between the diffusion tractography and axonal tracing of STP images. (D-G) The detected GFP signals (green) of vlPFC projectome and the IFOF tracts (red) obtained by diffusion tractography were overlaid on anatomical MRI images, with a magnified view of the box area. Evidently there was no fluorescent signal detected in the superior temporal area where the dMRI-derived IFOF tract passes through (G).
-

Evaluation Summary:
This paper will be of broad interest to readers who study anatomical connections of the brain. It demonstrates the efficacy of a cutting-edge viral tracing technique in mapping excitatory projections in macaque monkeys. The work describes the generation of a projectome from the macaque vlPFC cortex across the rest of the brain using AAV2/9-CaMKIIa-Tau-GFP labeling and imaging with high-throughput serial two-photon tomography. The comparison with imaging techniques available in humans (diffusion tractography) will also be of interest to research in human brain anatomy.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #1 agreed to share their name with the authors.)
-

Reviewer #1 (Public Review):
The authors report the generation of a mesoscale excitatory projectome from the ventrolateral prefrontal cortex (vlPFC) in the macaque brain by using AAV2/9-CaMKIIa-Tau-GFP labeling and imaging with high-throughput serial two-photon tomography. They present a novel data pipeline that integrates the STP data with macroscopic dMRI data from the same brain in a common 3D space, achieving a direct comparison of the two tracing methods. The analysis of the data revealed an interesting discrepancy between the high resolution STP data and the lower resolution dMRI data with respect to the extent of the frontal lobe projection through the inferior fronto-occipital fasciculus (IFOF) - the longest associative axon bundle in the human brain.
The authors report the generation of a mesoscale excitatory projectome from …
Reviewer #1 (Public Review):
The authors report the generation of a mesoscale excitatory projectome from the ventrolateral prefrontal cortex (vlPFC) in the macaque brain by using AAV2/9-CaMKIIa-Tau-GFP labeling and imaging with high-throughput serial two-photon tomography. They present a novel data pipeline that integrates the STP data with macroscopic dMRI data from the same brain in a common 3D space, achieving a direct comparison of the two tracing methods. The analysis of the data revealed an interesting discrepancy between the high resolution STP data and the lower resolution dMRI data with respect to the extent of the frontal lobe projection through the inferior fronto-occipital fasciculus (IFOF) - the longest associative axon bundle in the human brain.
The authors report the generation of a mesoscale excitatory projectome from the ventrolateral prefrontal cortex (vlPFC) in the macaque brain by using AAV2/9-CaMKIIa-Tau-GFP labeling and imaging with high-throughput serial two-photon tomography. They also present a novel data pipeline that integrates the STP data with macroscopic dMRI data from the same brain in a common 3D space, achieving a direct comparison of the two tracing methods. Overall the paper can serve as a how to example for analyzing large non-human primate brain data, though some parts of the paper can be improved and the interpretation of the data should also be further strengthened.
The methodological part should include more detail on image acquisition - speed of imaging, pixel residence time, total time for data acquisition of a single brain and data sizes. Also the time and hardware needed for the computational analysis should be included, including the registration to the common reference and the running time for the machine learning predictions - this should also include the F score for the axon detection.
The discrepancy between the high resolution STP data and the lower resolution dMRI data with respect to the extent of the frontal lobe projection through the inferior fronto-occipital fasciculus seems puzzling. One would expect that the STP data would reveal more detail not less.. One possibility is that the Tau-GFP does not diffuse throughout the full axon arborization of the PFC neurons, resulting in a technical artifact. Can this be excluded to support the functional significance of the current data?
-

Reviewer #2 (Public Review):
The authors utilized viral vectors as neural tracers to delineate the connectivity map of the macaque vlPFC at the axonal level. There are three main goals of this study: 1) determine an effective viral vector for tract-tracing in the macaque brain, 2) delineate the detailed map of excitatory vlPFC projections to the rest of the brain, and 3) compare vlPFC connectivity between tracing and tractography results.
Accordingly, my comments are organized around each aim:
- This study demonstrates the advantage of viral tracing technique in targeting neuron type-specific pathways. The authors conducted injection experiments with three types of viral vectors and found success of AAV in labeling long-distance connections without causing fatal neurotoxicity in the monkey. This success extends the application of AAV from …
Reviewer #2 (Public Review):
The authors utilized viral vectors as neural tracers to delineate the connectivity map of the macaque vlPFC at the axonal level. There are three main goals of this study: 1) determine an effective viral vector for tract-tracing in the macaque brain, 2) delineate the detailed map of excitatory vlPFC projections to the rest of the brain, and 3) compare vlPFC connectivity between tracing and tractography results.
Accordingly, my comments are organized around each aim:
- This study demonstrates the advantage of viral tracing technique in targeting neuron type-specific pathways. The authors conducted injection experiments with three types of viral vectors and found success of AAV in labeling long-distance connections without causing fatal neurotoxicity in the monkey. This success extends the application of AAV from rodents to nonhuman primates. The fact that AAV specifically targets glutamatergic neurons makes it advantageous for mapping excitatory projections.
Although the labeling efficacy of each viral vector type is described in the text, Fig. 2 does not present a clear comparison across viral vectors, despite such comparison for a thalamic injection in Fig. 2S. Without a comparable graph to Fig. 2E, it is unclear to what extent the VSV and lentivirus failed in labeling long-distance pathways.
- The authors quantified connectivity strength by the GFP signal intensity using a machine-learning algorithm. Both the quantitative approach and the resulting excitatory projection map are important contributions to advancing our knowledge of vlPFC connectivity.
However, several issues with the analysis lead to concerns about the connectivity result. First, the strength measure is based on axonal patterns in the terminal fields (which the authors refer to as "axon clusters"), detected by a machine-learning algorithm (page 25, lines 11-13). However, the actual synaptic connections are the small dot-looking signals in the background. These "green dots" are boutons on the dendritic trees. The density of boutons rather than the passing fibers reflects the density of synapses. The brief method description does not mention how the boutons are quantified, and it is unclear whether the signal was treated as the background noise and filtered out. Second, it is difficult for the reader to assess the robustness of the vlPFC connectivity patterns, due to these issues: i) It is unclear how many injection cases were used to generate the result reported in the subsection "Brain-wide excitatory projectome of vlPFC in macaques". The text mentions a singular "injection site" (page 8, line 12) and Fig. 4 shows a single site. However, there are three cases listed in Table 1. Is the result an average of all three cases? ii) Relatedly, it is unclear in which anatomical area the injection was placed for each case. Table 1 lists the site as "vlPFC" for all three cases, while the vlPFC contains areas 44, 45 and 12l. These areas have different projection patterns documented in the tract tracing literature. If different areas were injected in the three cases, they should be reported separately. iii) It is hard to compare the projection patterns with those reported in the literature. Conventionally, tract tracing studies report terminal fields by showing original labeling patterns in both cortical and subcortical regions without averaging within divided areas (see e.g. Petrides & Pandya, 2007, J Neurosci). It is hard to compare Fig. 3 with previous tract tracing studies to assess its robustness.
- Using the ground-truth from tract tracing to validate tractography results is a timely problem and this study showed promising consistency and discrepancy between the two modalities. Especially, the discrepancy between tracing and tractography data on the IFOF termination brings critical insights into a potential cross-species difference. The finding that IFOF does not reach the occipital cortex provides important support for the speculation that IFOF may not exist in monkeys (for a context of the IFOF debate see Schmahmann & Pandya, 2006, pp 445-446).
I have minor concerns regarding the statistical robustness of the tracing-tractography comparison. The authors compared the vlPFC-CC-contralateral tract instead of a global connectivity pattern without justification. Why omitting other major tracts that connect with vlPFC? In addition, the results are shown for only one monkey, while two monkeys went through both tracer injection and dMRI scans. It is unclear how the results were chosen or whether the data were averaged.
-