METTL18-mediated histidine methylation of RPL3 modulates translation elongation for proteostasis maintenance
Curation statements for this article:-
Curated by eLife
Evaluation Summary:
This work examines how METTLL18-mediated RPL3 histidine methylation on 245 position regulates translation elongation and protects cells from cellular aggregation of Tyr-rich proteins. The study hints at the existence of a "ribosome code" and how posttranslational modification of ribosomal proteins could affect translation.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #1 and Reviewer #3 agreed to share their names with the authors.)
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Protein methylation occurs predominantly on lysine and arginine residues, but histidine also serves as a methylation substrate. However, a limited number of enzymes responsible for this modification have been reported. Moreover, the biological role of histidine methylation has remained poorly understood to date. Here, we report that human METTL18 is a histidine methyltransferase for the ribosomal protein RPL3 and that the modification specifically slows ribosome traversal on Tyr codons, allowing the proper folding of synthesized proteins. By performing an in vitro methylation assay with a methyl donor analog and quantitative mass spectrometry, we found that His245 of RPL3 is methylated at the τ- N position by METTL18. Structural comparison of the modified and unmodified ribosomes showed stoichiometric modification and suggested a role in translation reactions. Indeed, genome-wide ribosome profiling and an in vitro translation assay revealed that translation elongation at Tyr codons was suppressed by RPL3 methylation. Because the slower elongation provides enough time for nascent protein folding, RPL3 methylation protects cells from the cellular aggregation of Tyr-rich proteins. Our results reveal histidine methylation as an example of a ribosome modification that ensures proteome integrity in cells.
Article activity feed
-

-

Author Response
Reviewer #1 (Public Review):
Overall, this study is well designed with convincing experimental data. The following critiques should be considered:
- It is important to examine whether the phenotype of METTL18 KO is mediated through change with RPL3 methylation. The functional link between METTL18 and RPL3 methylation on regulating translation elongation need to be examined in details.
We truly thank the reviewer for the suggestion. Accordingly, we set up experiments combined with hybrid in vitro translation (Panthu et al. Biochem J 2015 and Erales et al. PNAS 2017) and the Renilla–firefly luciferase fusion reporter system (Kisly et al. NAR 2021) (see Figure 5A).
To test the impact of RPL3 methylation on translation directly, we purified ribosomes from METTL18 KO cells or naïve HEK293T cells supplemented with …
Author Response
Reviewer #1 (Public Review):
Overall, this study is well designed with convincing experimental data. The following critiques should be considered:
- It is important to examine whether the phenotype of METTL18 KO is mediated through change with RPL3 methylation. The functional link between METTL18 and RPL3 methylation on regulating translation elongation need to be examined in details.
We truly thank the reviewer for the suggestion. Accordingly, we set up experiments combined with hybrid in vitro translation (Panthu et al. Biochem J 2015 and Erales et al. PNAS 2017) and the Renilla–firefly luciferase fusion reporter system (Kisly et al. NAR 2021) (see Figure 5A).
To test the impact of RPL3 methylation on translation directly, we purified ribosomes from METTL18 KO cells or naïve HEK293T cells supplemented with ribosome-depleted rabbit reticulocyte lysate (RRL) and then conducted an in vitro translation assay (i.e., hybrid translation, Panthu et al. Biochem J 2015 and Erales et al. PNAS 2017) (see figure above and Figure 5A). Indeed, we observed that removal of the ribosomes from RRL decreased protein synthesis in vitro and that the addition of ribosomes from HEK293T cells efficiently recovered the activity (see Figure 5 — figure supplement 1A).
To test the effect on Tyr codon elongation, we harnessed the fusion of Renilla and firefly luciferases; this system allows us to detect the delay/promotion of downstream firefly luciferase synthesis compared to upstream Renilla luciferase and thus to focus on elongation affected by the sequence inserted between the two luciferases (Kisly et al. NAR 2021) (see figure above and Figure 5A). For better detection of the effects on Tyr codons, we used the repeat of the codon (×39, the number was due to cloning constraints in our hands). We note that the insertion of Tyr codon repeats reduced the elongation rate (or processivity), as we observed a reduced slope of downstream Fluc synthesis (see Figure 5 — figure supplement 1B).
Using this setup, we observed that, compared to ribosomes from naïve cells, RPL3 methylation-deficient ribosomes led to faster elongation at Tyr repeats (see Figure 5B). These data, which are directly reflected by the ribosomes possessing unmethylated RPL3, provided solid evidence of a link between RPL3 methylation and translation elongation at Tyr codons.
- The obvious discrepancy between the recent NAR an this study lies in the ribosomal profiling results (such as Fig.S5). The cell line specific regulation between HAP1 (previously used in NAR) vs 293T cell used here ( in this study) needs to be explored. For example, would METLL18 KO in HAP1 cells cause polysome profiling difference in this study? Some of negative findings in this study (such as Fig.S3B, Fig.S5A) would need some kind of positive control to make sure that the assay condition would be working.
According to the reviewer’s suggestion, we conducted polysome profiling of the HAP1 cells with METTL18 knockout. For this assay, we used the same cell line (HAP1 METTL18 KO, 2-nt del.) as in the earlier NAR paper. As shown in Figure 9 — figure supplement 2A and 2B, we observed reduced polysomes in this cell line, as observed in the NAR paper.
We did not find the abundance of 40S and 60S by assessing the rRNAs and the complex mass in the sucrose gradient (see Figure 9 — figure supplement 2C-E) by METTL18 KO in HAP1 cells. This observation was again consistent with earlier reports.
Overall, our experiments in sucrose density gradient (polysome and 40S/60S ratio) were congruent with NAR paper. A difference from our finding in HEK293T cells was the limited effect on polysome formation by METTL18 deletion (Figure 4 — figure supplement 1A and 1B). To further provide a careful control for this observation, we induced a 60S biogenesis delay, as requested by the Reviewer. Here, we treated cells with siRNA targeting RPL17, which is needed for proper 60S assembly (Wang et al. RNA 2015). The quantification of SDG showed a reduction of 60S (see figure below and Figure 3 — figure supplement 1D-F) and polysomes (see Figure 4 — figure supplement 1C and 1D), highlighting the weaker effects of METTL18 depletion on 60S and polysome formation in HEK293T cells. We note that all the sucrose density gradient experiments were repeated 3 times, quantified, and statistically tested.
To further assess the difference between our data and those in the earlier NAR paper, we also performed ribosome profiling on 3 independent KO lines in HAP1 cells, including the one used in the NAR paper (METTL18 KO, 2-nt del.). Indeed, all METTL18 KO HAP1 cells showed a reduction in footprints on Tyr codons, as observed in HEK293 cells (see Figure 4H), and thus, there was a consistent effect of RPL3 methylation on elongation irrespective of the cell type. On the other hand, we could not find such a trend (see figure below) by reanalysis of the published data (Małecki et al. NAR 2021).
Thus far, we could not find the origin of the difference in ribosome profiling compared to the earlier paper. Culture conditions or other conditions may affect the data. Given that, we amended the discussion to cover the potential of context/situation-dependent effects on RPL3 methylation.
- For loss-of-function studies of METLL18, it will be beneficial to have a second sgRNA to KO METLL18 to solidify the conclusion.
We thank the reviewer for the constructive suggestion. Instead of screening additional METTL18 KO in HEK293T cells, we conducted additional ribosome profiling experiments in HAP1 cells with 3 independent KO lines. In addition to ensuring reproducibility, these experiments should assess whether our results are specific to the HEK293T cells that we mainly used. As mentioned above, even in the different cell lines, we observed faster elongation of the Tyr codon by METTL18 deficiency.
- In addition to loss-of-function studies for METLL18, gain-of-function studies for METLL18 would be helpful for making this study more convincing.
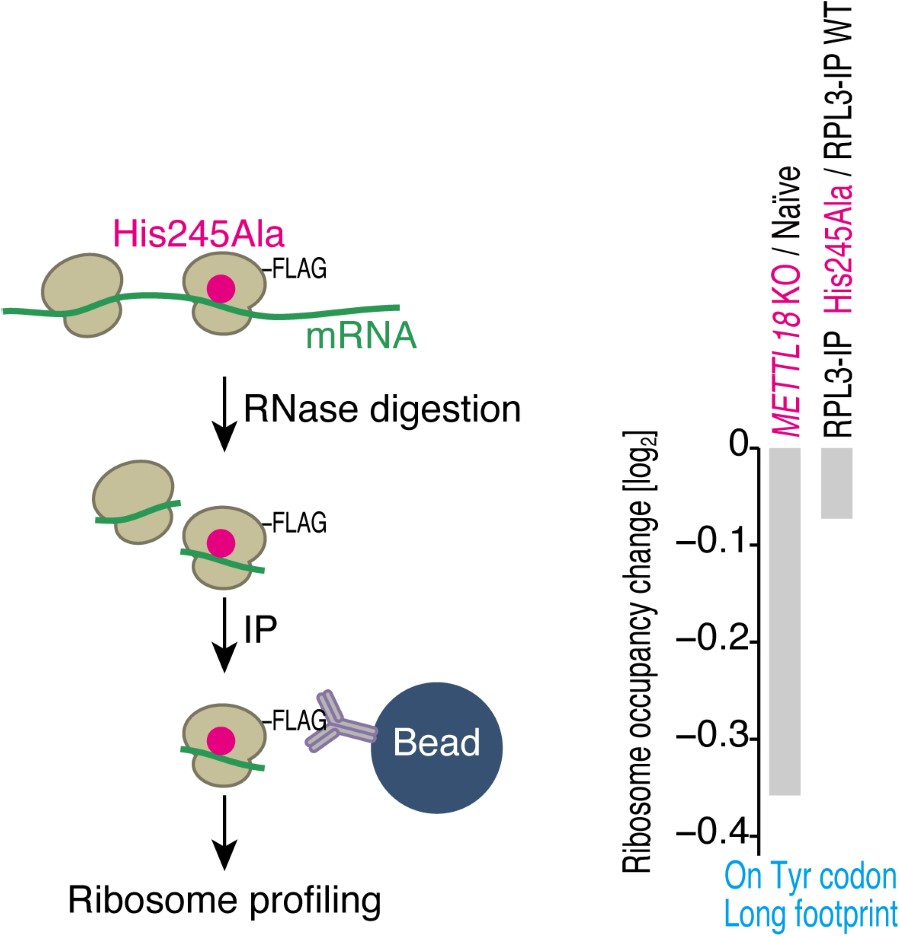
Again, we thank the reviewer for the constructive suggestion. To address this issue, we conducted RiboTag-IP and subsequent ribosome profiling. Here, we expressed Cterminal FLAG-tagged RPL3 of its WT and His245Ala mutant, in which METTL18 could not add methylation (Figure 2A), in HEK293T cells, treated the lysate with RNase, immunoprecipitated FLAG-tagged ribosomes, and then prepared a ribosome profiling library (see figure below, left). This experiment assessed the translation driven by the tagged ribosomes. Indeed, we observed that, compared to the difference in Tyr codon elongation in METTL18 KO vs. naïve cells, His245Ala provided weaker impacts (see figure below, right). Given that METTL18 KO provides unmodified His, the enhanced Tyr elongation may be mediated by the bare His but not by Ala in that position. Since this point may be beyond the scope of this study, we omitted it from the manuscript. However, we are happy to add the data to the supplementary figures if requested.
Reviewer #3 (Public Review):
In this article, Matsuura-Suzuki et al provided strong evidence that the mammalian protein METTL18 methylates a histidine residue in the ribosomal protein RPL3 using a combination of Click chemistry, quantitative mass spectrometry, and in vitro methylation assays. They showed that METTL18 was associated with early sucrose gradient fractions prior to the 40S peak on a polysome profile and interpreted that as evidence that RPL3 is modified early in the 60S subunit biogenesis pathway. They performed cryo-EM of ribosomes from a METTL18-knockout strain, and show that the methyl group on the histidine present in published cryo-EM data was missing in their new cryo-EM structure. The missing methyl group gave minor changes in the residue conformation, in keeping with the minor effects observed on translation. They performed ribosome profiling to determine what is being translated efficiently in cells with and without METTL18, and found decreased enrichment of Tyrosine codons in the A site of ribosomes from cells lacking METTL18. They further showed that longer ribosome footprints corresponding to sequences within ribosomes that have already bound to A-site tRNA contained less Tyrosine codons in the A site when lacking METTL18. This suggests methylation normally slows down elongation after tRNA loading but prior to EF-2 dissociation. They hypothesize that this decreased rate affects protein folding and follow up with fluorescence microscopy to show that EGFP aggregated more readily in cells lacking METTL18, suggesting that translation elongation slow down mediated by METTL18 leads to enhanced folding. Finally, they performed SILAC on aggregated proteins to confirm that more tyrosine was incorporated into protein aggregates from cells lacking METTL18.
The article is interesting and uses a large number of different techniques to present evidence that histidine methylation of RPL3 leads to decreased elongation rates at Tyrosine codons, allowing time for effective protein folding.
We thank the reviewer for the positive comments.
I agree with the interpretation of the results, although I do have minor concerns:
- The magnitude of each effect observed by ribosome profiling is very small, which is not unusual for ribosome modifications or methylation. Methylation seems to occur on all ribosomes in the cell since the modification is present in several cryo-EM structures. The authors suggest that the modification occurs during biogenesis prior to folding and being inaccessible to METTL18, so it is unlikely to be removed. For that reason, I do not think it is warranted to claim that this is an example of a ribosome code, or translation tuning. Those terms would indicate regulated modifications that come on and off of proteins, but the authors have not presented evidence that the activity is regulated (and don't really need to for this paper to be impactful).
We thank the reviewer for making this point, and we agree that the nuance of the wording may not fit our results. We amended the corresponding sentences to avoid using the terms “ribosome code” and “translation tuning” throughout the manuscript.
- In Figure 4-supplement 1, it appears there are slightly more 80S less 60S in the METTL18 knockout with no change in 40S. It might be normal variability in this cell type, but quantitation of the peaks from 2 or more experiments is needed to make the claim that ribosome biogenesis is unaffected by METTL18 deletion. Likewise, the authors need to quantitate the area under the curve for 40S and 60S levels from several replicates and show an average -/+ error for figure 3, supplement 1 because that result is essential to claim that ribosome biogenesis is unaffected.
Accordingly, we repeated all the sucrose density gradient experiments 3 times, quantified the data, and statistically tested the results. Even in the quantification, we could not find a significant change in either the 40S or 60S levels by METTL18 deletion in HEK293T cells (see Figure 3 — figure supplement 1B and 1C).
Moreover, for the positive control of 60S biogenesis delay, we treated cells with siRNA targeting RPL17, which is needed for proper 60S assembly (Wang et al. RNA 2015). The quantification of SDG showed a reduction in 60S (see figure below and Figure 3 — figure supplement 1D-F) and polysomes (see Figure 4 — figure supplement 1C and 1D), highlighting the weaker effects of METTL18 depletion on 60S and polysome formation.
- The effect of methylation could be any step after accommodation of tRNA in the A site and before dissociation of EF-2, including peptidyl transfer. More evidence is needed for claiming strongly that methylation slows translocation specifically. This could be followed up in vitro in a new study.
We truly thank the reviewer for the suggestion. Accordingly, we set up experiments combined with hybrid in vitro translation (Panthu et al. Biochem J 2015 and Erales et al. PNAS 2017) and the Renilla–firefly luciferase fusion reporter system (Kisly et al. NAR 2021) (see Figure 5A).
To test the impact of RPL3 methylation on translation directly, we purified ribosomes from METTL18 KO cells or naïve HEK293T cells supplemented with ribosome-depleted rabbit reticulocyte lysate (RRL) and then conducted an in vitro translation assay (i.e., hybrid translation, Panthu et al. Biochem J 2015 and Erales et al. PNAS 2017) (see figure above and Figure 5A). Indeed, we observed that removal of the ribosomes from RRL decreased protein synthesis in vitro and that the addition of ribosomes from HEK293T cells efficiently recovered the activity (see Figure 5 — figure supplement 1A).
To test the effect on Tyr codon elongation, we harnessed the fusion of Renilla and firefly luciferases; this system allows us to detect the delay/promotion of downstream firefly luciferase synthesis compared to upstream Renilla luciferase and thus to focus on elongation affected by the sequence inserted between the two luciferases (Kisly et al. NAR 2021) (see figure above and Figure 5A). For better detection of the effects on Tyr codons, we used the repeat of the codon (×39, the number was due to cloning constraints in our hands). We note that the insertion of Tyr codon repeats reduced the elongation rate (or processivity), as we observed a reduced slope of downstream Fluc synthesis (see Figure 5 — figure supplement 1B).
Using this setup, we observed that, compared to ribosomes from naïve cells, RPL3 methylation-deficient ribosomes led to faster elongation at Tyr repeats (see Figure 5B). These data, which are directly reflected by the ribosomes possessing unmethylated RPL3, provided solid evidence of a link between RPL3 methylation and translation elongation at Tyr codons.
-

Evaluation Summary:
This work examines how METTLL18-mediated RPL3 histidine methylation on 245 position regulates translation elongation and protects cells from cellular aggregation of Tyr-rich proteins. The study hints at the existence of a "ribosome code" and how posttranslational modification of ribosomal proteins could affect translation.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #1 and Reviewer #3 agreed to share their names with the authors.)
-

Reviewer #1 (Public Review):
Overall, this study is well designed with convincing experimental data. The following critiques should be considered:
1. It is important to examine whether the phenotype of METTL18 KO is mediated through change with RPL3 methylation. The functional link between METTL18 and RPL3 methylation on regulating translation elongation need to be examined in details.
2. The obvious discrepancy between the recent NAR an this study lies in the ribosomal profiling results (such as Fig.S5). The cell line specific regulation between HAP1 (previously used in NAR) vs 293T cell used here ( in this study) needs to be explored. For example, would METLL18 KO in HAP1 cells cause polysome profiling difference in this study? Some of negative findings in this study (such as Fig.S3B, Fig.S5A) would need some kind of positive control …
Reviewer #1 (Public Review):
Overall, this study is well designed with convincing experimental data. The following critiques should be considered:
1. It is important to examine whether the phenotype of METTL18 KO is mediated through change with RPL3 methylation. The functional link between METTL18 and RPL3 methylation on regulating translation elongation need to be examined in details.
2. The obvious discrepancy between the recent NAR an this study lies in the ribosomal profiling results (such as Fig.S5). The cell line specific regulation between HAP1 (previously used in NAR) vs 293T cell used here ( in this study) needs to be explored. For example, would METLL18 KO in HAP1 cells cause polysome profiling difference in this study? Some of negative findings in this study (such as Fig.S3B, Fig.S5A) would need some kind of positive control to make sure that the assay condition would be working.
3. For loss-of-function studies of METLL18, it will be beneficial to have a second sgRNA to KO METLL18 to solidify the conclusion.
4. In addition to loss-of-function studies for METLL18, gain-of-function studies for METLL18 would be helpful for making this study more convincing.
-

Reviewer #2 (Public Review):
The manuscript by Matsura-Suzuki et al. characterizes the role of METTL18 histidine methyltransferase in protein synthesis. The authors used genetical manipulation, affinity purification and mass spectrometry to indicate METTL18 protein as methyltransferase that specifically modifies ribosomal protein RPL3 during ribosome maturation. Using METTL18 based in vitro methylation system they annotate His245 residue in RPL3 protein as a methylhistidine and confirmed their results using a mass spectrometry on immunopurified RPL3 protein from HEK293 cells. The authors use ribosome profiling techniques, luciferase aggregation assays and mass spectrometry analyses of cellular aggregates to argue for Tyrosine specific effects during protein synthesis that influence protein folding.
While METTL18 was recently …Reviewer #2 (Public Review):
The manuscript by Matsura-Suzuki et al. characterizes the role of METTL18 histidine methyltransferase in protein synthesis. The authors used genetical manipulation, affinity purification and mass spectrometry to indicate METTL18 protein as methyltransferase that specifically modifies ribosomal protein RPL3 during ribosome maturation. Using METTL18 based in vitro methylation system they annotate His245 residue in RPL3 protein as a methylhistidine and confirmed their results using a mass spectrometry on immunopurified RPL3 protein from HEK293 cells. The authors use ribosome profiling techniques, luciferase aggregation assays and mass spectrometry analyses of cellular aggregates to argue for Tyrosine specific effects during protein synthesis that influence protein folding.
While METTL18 was recently characterized as a RPL3 specific histidine methyltransferase in HAP1 cells by another group (Malecki et al., 2021). The previously published study also pointed out on the role of this modification in ribosome biogenesis, general translation and GAA codon specific effects, however the study by Matsura-Suzuki et al., argues for Tyrosine specific effects and impact of RPL3 His245 modification on proteostasis maintenance which would clearly distinguish these two studies and importance of the METTL18. -

Reviewer #3 (Public Review):
In this article, Matsuura-Suzuki et al provided strong evidence that the mammalian protein METTL18 methylates a histidine residue in the ribosomal protein RPL3 using a combination of Click chemistry, quantitative mass spectrometry, and in vitro methylation assays. They showed that METTL18 was associated with early sucrose gradient fractions prior to the 40S peak on a polysome profile and interpreted that as evidence that RPL3 is modified early in the 60S subunit biogenesis pathway. They performed cryo-EM of ribosomes from a METTL18-knockout strain, and show that the methyl group on the histidine present in published cryo-EM data was missing in their new cryo-EM structure. The missing methyl group gave minor changes in the residue conformation, in keeping with the minor effects observed on translation. They …
Reviewer #3 (Public Review):
In this article, Matsuura-Suzuki et al provided strong evidence that the mammalian protein METTL18 methylates a histidine residue in the ribosomal protein RPL3 using a combination of Click chemistry, quantitative mass spectrometry, and in vitro methylation assays. They showed that METTL18 was associated with early sucrose gradient fractions prior to the 40S peak on a polysome profile and interpreted that as evidence that RPL3 is modified early in the 60S subunit biogenesis pathway. They performed cryo-EM of ribosomes from a METTL18-knockout strain, and show that the methyl group on the histidine present in published cryo-EM data was missing in their new cryo-EM structure. The missing methyl group gave minor changes in the residue conformation, in keeping with the minor effects observed on translation. They performed ribosome profiling to determine what is being translated efficiently in cells with and without METTL18, and found decreased enrichment of Tyrosine codons in the A site of ribosomes from cells lacking METTL18. They further showed that longer ribosome footprints corresponding to sequences within ribosomes that have already bound to A-site tRNA contained less Tyrosine codons in the A site when lacking METTL18. This suggests methylation normally slows down elongation after tRNA loading but prior to EF-2 dissociation. They hypothesize that this decreased rate affects protein folding and follow up with fluorescence microscopy to show that EGFP aggregated more readily in cells lacking METTL18, suggesting that translation elongation slow down mediated by METTL18 leads to enhanced folding. Finally, they performed SILAC on aggregated proteins to confirm that more tyrosine was incorporated into protein aggregates from cells lacking METTL18.
The article is interesting and uses a large number of different techniques to present evidence that histidine methylation of RPL3 leads to decreased elongation rates at Tyrosine codons, allowing time for effective protein folding. I agree with the interpretation of the results, although I do have minor concerns:
1. The magnitude of each effect observed by ribosome profiling is very small, which is not unusual for ribosome modifications or methylation. Methylation seems to occur on all ribosomes in the cell since the modification is present in several cryo-EM structures. The authors suggest that the modification occurs during biogenesis prior to folding and being inaccessible to METTL18, so it is unlikely to be removed. For that reason, I do not think it is warranted to claim that this is an example of a ribosome code, or translation tuning. Those terms would indicate regulated modifications that come on and off of proteins, but the authors have not presented evidence that the activity is regulated (and don't really need to for this paper to be impactful).
2. In Figure 4-supplement 1, it appears there are slightly more 80S less 60S in the METTL18 knockout with no change in 40S. It might be normal variability in this cell type, but quantitation of the peaks from 2 or more experiments is needed to make the claim that ribosome biogenesis is unaffected by METTL18 deletion. Likewise, the authors need to quantitate the area under the curve for 40S and 60S levels from several replicates and show an average -/+ error for figure 3, supplement 1 because that result is essential to claim that ribosome biogenesis is unaffected.
3. The effect of methylation could be any step after accommodation of tRNA in the A site and before dissociation of EF-2, including peptidyl transfer. More evidence is needed for claiming strongly that methylation slows translocation specifically. This could be followed up in vitro in a new study.
-