The BigBrainWarp toolbox for integration of BigBrain 3D histology with multimodal neuroimaging
Curation statements for this article:-
Curated by eLife
Evaluation Summary:
The manuscript introduces a new tool - BigBrainWarp - which consolidates several of the tools used to analyse BigBrain into a single, easy to use and well documented tool. The BigBrain project produced the first open, high-resolution cell-scale histological atlas of a whole human brain. The tool presented here should make it easy for any researcher to use the wealth of information available in the BigBrain for the annotation of their own neuroimaging data. This is an important resource, with diverse tutorials demonstrating broad application.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #1, Reviewer #2 and Reviewer #3 agreed to share their names with the authors.)
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Neuroimaging stands to benefit from emerging ultrahigh-resolution 3D histological atlases of the human brain; the first of which is ‘BigBrain’. Here, we review recent methodological advances for the integration of BigBrain with multi-modal neuroimaging and introduce a toolbox, ’BigBrainWarp’, that combines these developments. The aim of BigBrainWarp is to simplify workflows and support the adoption of best practices. This is accomplished with a simple wrapper function that allows users to easily map data between BigBrain and standard MRI spaces. The function automatically pulls specialised transformation procedures, based on ongoing research from a wide collaborative network of researchers. Additionally, the toolbox improves accessibility of histological information through dissemination of ready-to-use cytoarchitectural features. Finally, we demonstrate the utility of BigBrainWarp with three tutorials and discuss the potential of the toolbox to support multi-scale investigations of brain organisation.
Article activity feed
-

-

Author Response:
Reviewer #1:
- The user manual and tutorial are well documented, although the actual code could do with more explicit documentation and comments throughout. The overall organisation of the code is also a bit messy.
We have now implemented an ongoing, automated code review via Codacy (https://app.codacy.com/gh/caseypaquola/BigBrainWarp/dashboard). The grade is published as a badge on GitHub. We improved the quality of the code to an A grade by increasing comments and fixing code style issues. Additionally, we standardised the nomenclature throughout the toolbox to improve consistency across scripts and we restructured the bigbrainwarp function.
- My understanding is that this toolbox can take maps from BigBrain to MRI space and vice versa, but the maps that go in the direction BigBrain->MRI seem to be confined to those …
Author Response:
Reviewer #1:
- The user manual and tutorial are well documented, although the actual code could do with more explicit documentation and comments throughout. The overall organisation of the code is also a bit messy.
We have now implemented an ongoing, automated code review via Codacy (https://app.codacy.com/gh/caseypaquola/BigBrainWarp/dashboard). The grade is published as a badge on GitHub. We improved the quality of the code to an A grade by increasing comments and fixing code style issues. Additionally, we standardised the nomenclature throughout the toolbox to improve consistency across scripts and we restructured the bigbrainwarp function.
- My understanding is that this toolbox can take maps from BigBrain to MRI space and vice versa, but the maps that go in the direction BigBrain->MRI seem to be confined to those provided in the toolbox (essentially the density profiles). What if someone wants to do some different analysis on the BigBrain data (e.g. looking at cellular morphology) and wants that mapped onto MRI spaces? Does this tool allow for analyses that involve the raw BigBrain data? If so, then at what resolution and with what scripts? I think this tool will have much more impact if that was possible. Currently, it looks as though the 3 tutorial examples are basically the only thing that can be done (although I may be lacking imagination here).
The bigbrainwarp function allows input of raw BigBrain data in volume and surface forms. For volumetric inputs, the image must be aligned to the full BigBrain or BigBrainSym volume, but the function is agnostic to the input voxel resolution. We have also added an option for the user to specify the output voxel resolution. For example,
bigbrainwarp --in_space bigbrain --in_vol cellular_morphology_in_bigbrain.nii \ --interp linear --out_space icbm --out_res 0.5
--desc cellular_morphology --wd working_directorywhere “cellular_morphology_in_bigbrain.nii” was generated from a BigBrain volume (see Table 2 below for all parameters). The BigBrain volume may be the 100-1000um resolution images provided on the ftp or a resampled version of these images, as long as the full field of view is maintained. For surface-based inputs, the data must contain a value for each vertex of the BigBrain/BigBrainSym mesh. We have clarified these points in the Methods, illustrated the potential transformations in an extended Figure 3 and highlighted the distinctiveness of the tutorial transformations in the Results.
- An obvious caveat to bigbrain is that it is a single brain and we know there are sometimes substantial individual variations in e.g. areal definition. This is only slightly touched upon in the discussion. Might be worth commenting on this more. As I see it, there are multiple considerations. For example (i) Surface-to-Surface registration in the presence of morphological idiosyncracies: what parts of the brain can we "trust" and what parts are uncertain? (ii) MRI parcellations mapped onto BigBrain will vary in how accurately they may reflect the BigBrain areal boundaries: if histo boundaries do not correspond with MRI-derived ones, is that because BigBrain is slightly different or is it a genuine divergence between modalities? Of course addressing these questions is out of scope of this manuscript, but some discussion could be useful; I also think this toolbox may be useful for addressing this very concerns!
We agree that these are important questions and hope that BigBrainWarp will propel further research. Here, we consider these questions from two perspectives; the accuracy of the transformations and the potential influence of individual variation. For the former, we conducted a quantitative analysis on the accuracy of transformations used in BigBrainWarp (new Figure 2). We provide a function (evaluate_warp.sh) for BigBrainWarp users to assess accuracy of novel deformation fields and encourage detailed inspection of accuracy estimates and deformation effects for region of interest studies. For the latter, we expanded our Discussion of previous research on inter-individual variability and comment on the potential implications of unquantified inter-individual variability for the interpretation of BigBrain-MRI comparisons.
Methods (P.7-8):
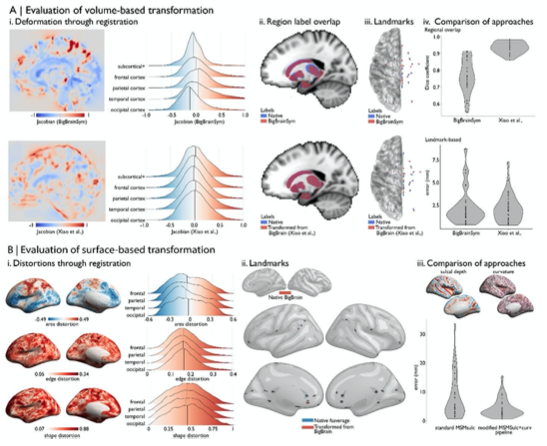
“A prior study (Xiao et al., 2019) was able to further improve the accuracy of the transformation for subcortical structures and the hippocampus using a two-stage multi-contrast registration. The first stage involved nonlinear registration of BigBrainSym to a PD25 T1-T2* fusion atlas (Xiao et al., 2017, 2015), using manual segmentations of the basal ganglia, red nucleus, thalamus, amygdala, and hippocampus as additional shape priors. Notably, the PD25 T1-T2* fusion contrast is more similar to the BigBrainSym intensity contrast than a T1-weighted image. The second stage involved nonlinear registration of PD25 to ICBM2009sym and ICBM2009asym using multiple contrasts. The deformation fields were made available on Open Science Framework (https://osf.io/xkqb3/). The accuracy of the transformations was evaluated relative to overlap of region labels and alignment of anatomical fiducials (Lau et al., 2019). The two-stage procedure resulted in 0.86-0.97 Dice coefficients for region labels, improving upon direct overlap of BigBrainSym with ICBM2009sym (0.55-0.91 Dice) (Figure 2Aii, 2Aiv top). Transformed anatomical fiducials exhibited 1.77±1.25mm errors, on par with direct overlap of BigBrainSym with ICBM2009sym (1.83±1.47mm) (Figure 2Aiii, 2Aiv below). The maximum misregistration distance (BigBrainSym=6.36mm, Xiao=5.29mm) provides an approximation of the degree of uncertainty in the transformation. In line with this work, BigBrainWarp enables evaluation of novel deformation fields using anatomical fiducials and region labels (evaluate_warps.sh). The script accepts a nonlinear transformation file for registration of BigBrainSym to ICBM2009sym, or vice versa, and returns the Jacobian map, Dice coefficients for labelled regions and landmark misregistration distances for the anatomical fiducials.
The unique morphology of BigBrain also presents challenges for surface-based transformations. Idiosyncratic gyrification of certain regions of BigBrain, especially the anterior cingulate, cause misregistration (Lewis et al., 2020). Additionally, the areal midline representation of BigBrain, following inflation to a sphere, is disproportionately smaller than standard surface templates, which is related to differences in surface area, in hemisphere separation methods, and in tessellation methods. To overcome these issues, ongoing work (Lewis et al., 2020) combines a specialised BigBrain surface mesh with multimodal surface matching [MSM; (Robinson et al., 2018, 2014)] to co-register BigBrain to standard surface templates. In the first step, the BigBrain surface meshes were re-tessellated as unstructured meshes with variable vertex density (Möbius and Kobbelt, 2010) to be more compatible with FreeSurfer generated meshes. Then, coarse-to-fine MSM registration was applied in three stages. An affine rotation was applied to the BigBrain sphere, with an additional “nudge” based on an anterior cingulate landmark. Next, nonlinear/discrete alignment using sulcal depth maps (emphasising global scale, Figure 2Biii), followed by nonlinear/discrete alignment using curvature maps (emphasising finer detail, Figure 2Biii). The higher- order MSM procedure that was implemented for BigBrain maximises concordance of these features while minimising surface deformations in a physically plausible manner, accounting for size and shape distortions (Figure 2Bi) (Knutsen et al., 2010; Robinson et al., 2018). This modified MSMsulc+curv pipeline improves the accuracy of transformed cortical maps (4.38±3.25mm), compared to a standard MSMsulc approach (8.02±7.53mm) (Figure 2Bii-iii) (Lewis et al., 2020).”
Figure 2: Evaluating BigBrain-MRI transformations. A) Volume-based transformations i. Jacobian determinant of deformation field shown with a sagittal slice and stratified by lobe. Subcortical+ includes the shape priors (as described in Methods) and the + connotes hippocampus, which is allocortical. Lobe labels were defined based on assignment of CerebrA atlas labels (Manera et al., 2020) to each lobe. ii. Sagittal slices illustrate the overlap of native ICBM2009b and transformed subcortical+ labels. iii. Superior view of anatomical fiducials (Lau et al., 2019). iv. Violin plots show the DICE coefficient of regional overlap (ii) and landmark misregistration (iii) for the BigBrainSym and Xiao et al., approaches. Higher DICE coefficients shown improved registration of subcortical+ regions with Xiao et al., while distributions of landmark misregistration indicate similar performance for alignment of anatomical fiducials. B) Surface-based transformations. i. Inflated BigBrain surface projections and ridgeplots illustrate regional variation in the distortions of the mesh invoked by the modified MSMsulc+curv pipeline. ii. Eighteen anatomical landmarks shown on the inflated BigBrain surface (above) and inflated fsaverage (below). BigBrain landmarks were transformed to fsaverage using the modified MSMsulc+curv pipeline. Accuracy of the transformation was calculated on fsaverage as the geodesic distance between landmarks transformed from BigBrain and the native fsaverage landmarks. iii. Sulcal depth and curvature maps are shown on inflated BigBrain surface. Violin plots show the improved accuracy of the transformation using the modified MSMsulc+curv pipeline, compared to a standard MSMsulc approach.
Discussion (P.18):
“Cortical folding is variably associated with cytoarchitecture, however. The correspondence of morphology with cytoarchitectonic boundaries is stronger in primary sensory than association cortex (Fischl et al., 2008; Rajkowska and Goldman-Rakic, 1995a, 1995b). Incorporating more anatomical information in the alignment algorithm, such as intracortical myelin or connectivity, may benefit registration, as has been shown in neuroimaging (Orasanu et al., 2016; Robinson et al., 2018; Tardif et al., 2015). Overall, evaluating the accuracy of volume- and surface-based transformations is important for selecting the optimal procedure given a specific research question and to gauge the degree of uncertainty in a registration.”
Discussion (P.19):
“Despite all its promises, the singular nature of BigBrain currently prohibits replication and does not capture important inter-individual variation. While large-scale cytoarchitectural patterns are conserved across individuals, the position of areal boundaries relative to sulci vary, especially in association cortex (Amunts et al., 2020; Fischl et al., 2008; Zilles and Amunts, 2013) . This can affect interpretation of BigBrain-MRI comparisons. For instance, in tutorial 3, low predictive accuracy of functional communities by cytoarchitecture may be attributable to the subject- specific topographies, which are well established in functional imaging (Benkarim et al., 2020; Braga and Buckner, 2017; Gordon et al., 2017; Kong et al., 2019). Future studies should consider the influence of inter-subject variability in concert with the precision of transformations, as these two elements of uncertainty can impact our interpretations, especially at higher granularity.”
Reviewer #2:
This is a nice paper presenting a review of recent developments and research resulting from BigBrain and a tutorial guiding use of the BigBrainWarp toolbox. This toolbox supports registration to, and from, standard MRI volumetric and surface templates, together with mapping derived features between spaces. Examples include projecting histological gradients estimated from BigBrain onto fsaverage (and the ICMB2009 atlas) and projecting Yeo functional parcels onto the BigBrain atlas.
The key strength of this paper is that it supports and expands on a comprehensive tutorial and docker support available from the website. The tutorials there go into even more detail (with accompanying bash scripts) of how to run the full pipelines detailed in the paper. The docker makes the tool very easy to install but I was also able to install from source. The tutorials are diverse examples of broad possible applications; as such the combined resource has the potential to be highly impactful.
The minor weaknesses of the paper relate to its clarity and depth. Firstly, I found the motivations of the paper initially unclear from the abstract. I would recommend much more clearly stating that this is a review paper of recent research developments resulting from the BigBrain atlas, and a tutorial to accompany the bash scripts which apply the warps between spaces. The registration methodology is explained elsewhere.
In the revised Abstract (P.1), we emphasise that the manuscript involves a review of recent literature, the introduction of BigBrainWarp, and easy-to-follow tutorials to demonstrate its utility.
“Neuroimaging stands to benefit from emerging ultrahigh-resolution 3D histological atlases of the human brain; the first of which is “BigBrain”. Here, we review recent methodological advances for the integration of BigBrain with multi-modal neuroimaging and introduce a toolbox, “BigBrainWarp", that combines these developments. The aim of BigBrainWarp is to simplify workflows and support the adoption of best practices. This is accomplished with a simple wrapper function that allows users to easily map data between BigBrain and standard MRI spaces. The function automatically pulls specialised transformation procedures, based on ongoing research from a wide collaborative network of researchers. Additionally, the toolbox improves accessibility of histological information through dissemination of ready-to-use cytoarchitectural features. Finally, we demonstrate the utility of BigBrainWarp with three tutorials and discuss the potential of the toolbox to support multi-scale investigations of brain organisation.”
I also found parts of the paper difficult to follow - as a methodologist without comprehensive neuroanatomical terminology, I would recommend the review of past work to be written in a more 'lay' way. In many cases, the figure captions also seemed insufficient at first. For example it was not immediately obvious to me what is meant by 'mesiotemporal confluence' and Fig 1G is not referenced specifically in the text. In Fig 3C it is not immediately clear from the text of the caption that the cortical image is representing the correlation from the plots - specifically since functional connectivity is itself estimated through correlation.
In the updated manuscript, we have tried to remove neuroanatomical jargon and clearly define uncommon terms at the first instance in text. For example,
“Evidence has been provided that cortical organisation goes beyond a segregation into areas. For example, large- scale gradients that span areas and cytoarchitectonic heterogeneity within a cortical area have been reported (Amunts and Zilles, 2015; Goulas et al., 2018; Wang, 2020). Such progress became feasible through integration of classical techniques with computational methods, supporting more observer-independent evaluation of architectonic principles (Amunts et al., 2020; Paquola et al., 2019; Schiffer et al., 2020; Spitzer et al., 2018). This paves the way for novel investigations of the cellular landscape of the brain.”
“Using the proximal-distal axis of the hippocampus, we were able to bridge the isocortical and hippocampal surface models recapitulating the smooth confluence of cortical types in the mesiotemporal lobe, i.e. the mesiotemporal confluence (Figure 1G).”
“Here, we illustrate how we can track resting-state functional connectivity changes along the latero-medial axis of the mesiotemporal lobe, from parahippocampal isocortex towards hippocampal allocortex, hereafter referred to as the iso-to-allocortical axis.”
Additionally, we have expanded the captions for clarity. For example, Figure 3:
“C) Intrinsic functional connectivity was calculated between each voxel of the iso-to-allocortical axis and 1000 isocortical parcels. For each parcel, we calculated the product-moment correlation (r) of rsFC strength with iso-to- allocortical axis position. Thus, positive values (red) indicate that rsFC of that isocortical parcel with the mesiotemporal lobe increases along the iso-to-allocortex axis, whereas negative values (blue) indicate decrease in rsFC along the iso-to-allocortex axis.”
My minor concern is over the lack of details in relation to the registration pipelines. I understand these are either covered in previous papers or are probably destined for bespoke publications (in the case of the surface registration approach) but these details are important for readers to understand the constraints and limitations of the software. At this time, the details for the surface registration only relate to an OHBM poster and not a publication, which I was unable to find online until I went through the tutorial on the BigBrain website. In general I think a paper should have enough information on key techniques to stand alone without having to reference other publications, so, in my opinion, a high level review of these pipelines should be added here.
There isn't enough details on the registration. For the surface, what features were used to drive alignment, how was it parameterised (in particular the regularisation - strain, pairwise or areal), how was it pre-processed prior to running MSM - all these details seem to be in the excellent poster. I appreciate that work deserves a stand alone publication but some details are required here for users to understand the challenges, constraints and limitations of the alignment. Similar high level details should be given for the registration work.
We expanded descriptions of the registration strategies behind BigBrainWarp, especially so for the surface-based registration. Additionally, we created a new Figure to illustrate how the accuracy of the transformations may be evaluated.
Methods (P.7-8):
“For the initial BigBrain release (Amunts et al., 2013), full BigBrain volumes were resampled to ICBM2009sym (a symmetric MNI152 template) and MNI-ADNI (an older adult T1-weighted template) (Fonov et al., 2011). Registration of BigBrain to ICBM2009sym, known as BigBrainSym, involved a linear then a nonlinear transformation (available on ftp://bigbrain.loris.ca/BigBrainRelease.2015/). The nonlinear transformation was defined by a symmetric diffeomorphic optimiser [SyN algorithm, (Avants et al., 2008)] that maximised the cross- correlation of the BigBrain volume with inverted intensities and a population-averaged T1-weighted map in ICBM2009sym space. The Jacobian determinant of the deformation field illustrates the degree and direction of distortions on the BigBrain volume (Figure 2Ai top).
A prior study (Xiao et al., 2019) was able to further improve the accuracy of the transformation for subcortical structures and the hippocampus using a two-stage multi-contrast registration. The first stage involved nonlinear registration of BigBrainSym to a PD25 T1-T2* fusion atlas (Xiao et al., 2017, 2015), using manual segmentations of the basal ganglia, red nucleus, thalamus, amygdala, and hippocampus as additional shape priors. Notably, the PD25 T1-T2* fusion contrast is more similar to the BigBrainSym intensity contrast than a T1-weighted image. The second stage involved nonlinear registration of PD25 to ICBM2009sym and ICBM2009asym using multiple contrasts. The deformation fields were made available on Open Science Framework (https://osf.io/xkqb3/). The accuracy of the transformations was evaluated relative to overlap of region labels and alignment of anatomical fiducials (Lau et al., 2019). The two-stage procedure resulted in 0.86-0.97 Dice coefficients for region labels, improving upon direct overlap of BigBrainSym with ICBM2009sym (0.55-0.91 Dice) (Figure 2Aii, 2Aiv top). Transformed anatomical fiducials exhibited 1.77±1.25mm errors, on par with direct overlap of BigBrainSym with ICBM2009sym (1.83±1.47mm) (Figure 2Aiii, 2Aiv below). The maximum misregistration distance (BigBrainSym=6.36mm, Xiao=5.29mm) provides an approximation of the degree of uncertainty in the transformation. In line with this work, BigBrainWarp enables evaluation of novel deformation fields using anatomical fiducials and region labels (evaluate_warps.sh). The script accepts a nonlinear transformation file for registration of BigBrainSym to ICBM2009sym, or vice versa, and returns the Jacobian map, DICE coefficients for labelled regions and landmark misregistration distances for the anatomical fiducials.
The unique morphology of BigBrain also presents challenges for surface-based transformations. Idiosyncratic gyrification of certain regions of BigBrain, especially the anterior cingulate, cause misregistration (Lewis et al., 2020). Additionally, the areal midline representation of BigBrain, following inflation to a sphere, is disproportionately smaller than standard surface templates, which is related to differences in surface area, in hemisphere separation methods, and in tessellation methods. To overcome these issues, ongoing work (Lewis et al., 2020) combines a specialised BigBrain surface mesh with multimodal surface matching [MSM; (Robinson et al., 2018, 2014)] to co-register BigBrain to standard surface templates. In the first step, the BigBrain surface meshes were re-tessellated as unstructured meshes with variable vertex density (Möbius and Kobbelt, 2010) to be more compatible with FreeSurfer generated meshes. Then, coarse-to-fine MSM registration was applied in three stages. An affine rotation was applied to the BigBrain sphere, with an additional “nudge” based on an anterior cingulate landmark. Next, nonlinear/discrete alignment using sulcal depth maps (emphasising global scale, Figure 2Biii), followed by nonlinear/discrete alignment using curvature maps (emphasising finer detail, Figure 2Biii). The higher- order MSM procedure that was implemented for BigBrain maximises concordance of these features while minimising surface deformations in a physically plausible manner, accounting for size and shape distortions (Figure 2Bi) (Knutsen et al., 2010; Robinson et al., 2018). This modified MSMsulc+curv pipeline improves the accuracy of transformed cortical maps (4.38±3.25mm), compared to a standard MSMsulc approach (8.02±7.53mm) (Figure 2Bii-iii) (Lewis et al., 2020).”
(SEE FIGURE 2 in Response to Reviewer #1)
I would also recommend more guidance in terms of limitations relating to inter-subject variation. My interpretation of the results of tutorial 3, is that topographic variation of the cortex could easily be driving the greater variation of the frontal parietal networks. Either that, or the Yeo parcel has insufficient granularity; however, in that case any attempt to go to finer MRI driven parcellations - for example to the HCP parcellation, would create its own problems due to subject specific variability.
We agree that inter-individual variation may contribute to the low predictive accuracy of functional communities by cytoarchitecture. We expanded upon this possibility in the revised Discussion (P. 19) and recommend that future studies examine the uncertainty of subject-specific topographies in concert with uncertainties of transformations.
“These features depict the vast cytoarchitectural heterogeneity of the cortex and enable evaluation of homogeneity within imaging-based parcellations, for example macroscale functional communities (Yeo et al., 2011). The present analysis showed limited predictability of functional communities by cytoarchitectural profiles, even when accounting for uncertainty at the boundaries (Gordon et al., 2016). [...] Despite all its promises, the singular nature of BigBrain currently prohibits replication and does not capture important inter-individual variation. While large- scale cytoarchitectural patterns are conserved across individuals, the position of boundaries relative to sulci vary, especially in association cortex (Amunts et al., 2020; Fischl et al., 2008; Zilles and Amunts, 2013) . This can affect interpretation of BigBrain-MRI comparisons. For instance, in tutorial 3, low predictive accuracy of functional communities by cytoarchitecture may be attributable to the subject-specific topographies, which are well established in functional imaging (Benkarim et al., 2020; Braga and Buckner, 2017; Gordon et al., 2017; Kong et al., 2019). Future studies should consider the influence of inter-subject variability in concert with the precision of transformations, as these two elements of uncertainty can impact our interpretations, especially at higher granularity.”
Reviewer #3:
The authors make a point for the importance of considering high-resolution, cell-scale, histological knowledge for the analysis and interpretation of low-resolution MRI data. The manuscript describes the aims and relevance of the BigBrain project. The BigBrain is the whole brain of a single individual, sliced at 20µ and scanned at 1µ resolution. During the last years, a sustained work by the BigBrain team has led to the creation of a precise cell-scale, 3D reconstruction of this brain, together with manual and automatic segmentations of different structures. The manuscript introduces a new tool - BigBrainWarp - which consolidates several of the tools used to analyse BigBrain into a single, easy to use and well documented tool. This tool should make it easy for any researcher to use the wealth of information available in the BigBrain for the annotation of their own neuroimaging data. The authors provide three examples of utilisation of BigBrainWarp, and show the way in which this can provide additional insight for analysing and understanding neuroimaging data. The BigBrainWarp tool should have an important impact for neuroimaging research, helping bridge the multi-scale resolution gap, and providing a way for neuroimaging researchers to include cell-scale phenomena in their study of brain data. All data and code are available open source, open access.
Main concern:
One of the longstanding debates in the neuroimaging community concerns the relationship between brain geometry (in particular gyro/sulcal anatomy) and the cytoarchitectonic, connective and functional organisation of the brain. There are various examples of correspondance, but also many analyses showing its absence, particularly in associative cortex (for example, Fischl et al (2008) by some of the co-authors of the present manuscript). The manuscript emphasises the accuracy of their transformations to the different atlas spaces, which may give some readers a false impression. True: towards the end of the manuscript the authors briefly indicate the difficulty of having a single brain as source of histological data. I think, however, that the manuscript would benefit from making this point more clearly, providing the future users of BigBrainWarp with some conceptual elements and references that may help them properly apprise their results. In particular, it would be helpful to briefly describe which aspects of brain organisation where used to lead the deformation to the different templates, if they were only based on external anatomy, or if they took into account some other aspects such as myelination, thickness, …
We agree with the Reviewer that the accuracy of the transformation and the potential influence of inter-individual variability should be carefully considered in BigBrain-MRI studies. To highlight these issues in the updated manuscript, we first conducted a quantitative analysis on the accuracy of transformations used in BigBrainWarp (new Figure 2). We provide a function (evaluate_warp.sh) for users to assess accuracy of novel deformation fields and encourage detailed inspection of accuracy estimates and deformation effects for region of interest studies. Second, we expanded our discussion of previous research on inter-individual variability and comment on the potential implications of unquantified inter-individual variability for the interpretation of BigBrain-MRI comparisons.
Methods (P.7-8):
“A prior study (Xiao et al., 2019) was able to further improve the accuracy of the transformation for subcortical structures and the hippocampus using a two-stage multi-contrast registration. The first stage involved nonlinear registration of BigBrainSym to a PD25 T1-T2* fusion atlas (Xiao et al., 2017, 2015), using manual segmentations of the basal ganglia, red nucleus, thalamus, amygdala, and hippocampus as additional shape priors. Notably, the PD25 T1-T2* fusion contrast is more similar to the BigBrainSym intensity contrast than a T1-weighted image. The second stage involved nonlinear registration of PD25 to ICBM2009sym and ICBM2009asym using multiple contrasts. The deformation fields were made available on Open Science Framework (https://osf.io/xkqb3/). The accuracy of the transformations was evaluated relative to overlap of region labels and alignment of anatomical fiducials (Lau et al., 2019). The two-stage procedure resulted in 0.86-0.97 Dice coefficients for region labels, improving upon direct overlap of BigBrainSym with ICBM2009sym (0.55-0.91 Dice) (Figure 2Aii, 2Aiv top). Transformed anatomical fiducials exhibited 1.77±1.25mm errors, on par with direct overlap of BigBrainSym with ICBM2009sym (1.83±1.47mm) (Figure 2Aiii, 2Aiv below). The maximum misregistration distance (BigBrainSym=6.36mm, Xiao=5.29mm) provides an approximation of the degree of uncertainty in the transformation. In line with this work, BigBrainWarp enables evaluation of novel deformation fields using anatomical fiducials and region labels (evaluate_warps.sh). The script accepts a nonlinear transformation file for registration of BigBrainSym to ICBM2009sym, or vice versa, and returns the Jacobian map, Dice coefficients for labelled regions and landmark misregistration distances for the anatomical fiducials.
The unique morphology of BigBrain also presents challenges for surface-based transformations. Idiosyncratic gyrification of certain regions of BigBrain, especially the anterior cingulate, cause misregistration (Lewis et al., 2020). Additionally, the areal midline representation of BigBrain, following inflation to a sphere, is disproportionately smaller than standard surface templates, which is related to differences in surface area, in hemisphere separation methods, and in tessellation methods. To overcome these issues, ongoing work (Lewis et al., 2020) combines a specialised BigBrain surface mesh with multimodal surface matching [MSM; (Robinson et al., 2018, 2014)] to co-register BigBrain to standard surface templates. In the first step, the BigBrain surface meshes were re-tessellated as unstructured meshes with variable vertex density (Möbius and Kobbelt, 2010) to be more compatible with FreeSurfer generated meshes. Then, coarse-to-fine MSM registration was applied in three stages. An affine rotation was applied to the BigBrain sphere, with an additional “nudge” based on an anterior cingulate landmark. Next, nonlinear/discrete alignment using sulcal depth maps (emphasising global scale, Figure 2Biii), followed by nonlinear/discrete alignment using curvature maps (emphasising finer detail, Figure 2Biii). The higher- order MSM procedure that was implemented for BigBrain maximises concordance of these features while minimising surface deformations in a physically plausible manner, accounting for size and shape distortions (Figure 2Bi) (Knutsen et al., 2010; Robinson et al., 2018). This modified MSMsulc+curv pipeline improves the accuracy of transformed cortical maps (4.38±3.25mm), compared to a standard MSMsulc approach (8.02±7.53mm) (Figure 2Bii-iii) (Lewis et al., 2020).”
(SEE Figure 2 in response to previous reviewers)
Discussion (P.18, 19):
“Cortical folding is variably associated with cytoarchitecture, however. The correspondence of morphology with cytoarchitectonic boundaries is stronger in primary sensory than association cortex (Fischl et al., 2008; Rajkowska and Goldman-Rakic, 1995a, 1995b). Incorporating more anatomical information in the alignment algorithm, such as intracortical myelin or connectivity, may benefit registration, as has been shown in neuroimaging (Orasanu et al., 2016; Robinson et al., 2018; Tardif et al., 2015). Overall, evaluating the accuracy of volume- and surface-based transformations is important for selecting the optimal procedure given a specific research question and to gauge the degree of uncertainty in a registration.”
“Despite all its promises, the singular nature of BigBrain currently prohibits replication and does not capture important inter-individual variation. While large-scale cytoarchitectural patterns are conserved across individuals, the position of boundaries relative to sulci vary, especially in association cortex (Amunts et al., 2020; Fischl et al., 2008; Zilles and Amunts, 2013) . This can have implications on interpretation of BigBrain-MRI comparisons. For instance, in tutorial 3, low predictive accuracy of functional communities by cytoarchitecture may be attributable to the subject-specific topographies, which are well established in functional imaging (Benkarim et al., 2020; Braga and Buckner, 2017; Gordon et al., 2017; Kong et al., 2019). Future studies should consider the influence of inter- subject variability in concert with the precision of transformations, as these two elements of uncertainty can impact our interpretations, especially at higher granularity.”
Minor:
- In the abstract and later in p9 the authors talk about "state-of-the-art" non-linear deformation matrices. This may be confusing for some readers. To me, in brain imaging a matrix is most often a 4x4 affine matrix describing a linear transformation. However, the authors seem to be describing a more complex, non-linear deformation field. Whereas building a deformation matrix (4x4 affine) is not a big challenge, I agree that more sophisticated tools should provide more sophisticated deformation fields. The authors may consider using "deformation field" instead of "deformation matrix", but I leave that to their judgment.
As suggested, we changed the text to “deformation field” where relevant.
- In the results section, p11, the authors highlight the challenge of segmenting thalamic nuclei or different hippocampal regions, and suggest that this should be simplified by the use of the histological BigBrain data. However, the atlases currently provided in the OSF project do not include these more refined parcellation: there's one single "Thalamus" label, and one single "Hippocampus" label (not really single: left and right). This could be explicitly stated to prevent readers from having too high expectations (although I am certain that those finer parcellations should come in the very close future).
We updated the text to reflect the current state of such parcellations. While subthalamic nuclei are not yet segmented (to our knowledge), one of the present authors has segmented hippocampal subfields (https://osf.io/bqus3/) and we highlight this in the Results (P.11-12):
“Despite MRI acquisitions at high and ultra-high fields reaching submillimeter resolutions with ongoing technical advances, certain brain structures and subregions remain difficult to identify (Kulaga-Yoskovitz et al., 2015; Wisse et al., 2017; Yushkevich et al., 2015). For example, there are challenges in reliably defining the subthalamic nucleus (not yet released for BigBrain) or hippocampal Cornu Ammonis subfields [manual segmentation available on BigBrain, https://osf.io/bqus3/, (DeKraker et al., 2019)]. BigBrain-defined labels can be transformed to a standard imaging space for further investigation. Thus, this approach can support exploration of the functional architecture of histologically-defined regions of interest.”
-

Evaluation Summary:
The manuscript introduces a new tool - BigBrainWarp - which consolidates several of the tools used to analyse BigBrain into a single, easy to use and well documented tool. The BigBrain project produced the first open, high-resolution cell-scale histological atlas of a whole human brain. The tool presented here should make it easy for any researcher to use the wealth of information available in the BigBrain for the annotation of their own neuroimaging data. This is an important resource, with diverse tutorials demonstrating broad application.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #1, Reviewer #2 and Reviewer #3 agreed to share their names with the authors.)
-

Reviewer #1 (Public Review):
The user manual and tutorial are well documented, although the actual code could do with more explicit documentation and comments throughout. The overall organisation of the code is also a bit messy.
My understanding is that this toolbox can take maps from BigBrain to MRI space and vice versa, but the maps that go in the direction BigBrain->MRI seem to be confined to those provided in the toolbox (essentially the density profiles). What if someone wants to do some different analysis on the BigBrain data (e.g. looking at cellular morphology) and wants that mapped onto MRI spaces? Does this tool allow for analyses that involve the raw BigBrain data? If so, then at what resolution and with what scripts? I think this tool will have much more impact if that was possible. Currently, it looks as though the 3 …
Reviewer #1 (Public Review):
The user manual and tutorial are well documented, although the actual code could do with more explicit documentation and comments throughout. The overall organisation of the code is also a bit messy.
My understanding is that this toolbox can take maps from BigBrain to MRI space and vice versa, but the maps that go in the direction BigBrain->MRI seem to be confined to those provided in the toolbox (essentially the density profiles). What if someone wants to do some different analysis on the BigBrain data (e.g. looking at cellular morphology) and wants that mapped onto MRI spaces? Does this tool allow for analyses that involve the raw BigBrain data? If so, then at what resolution and with what scripts? I think this tool will have much more impact if that was possible. Currently, it looks as though the 3 tutorial examples are basically the only thing that can be done (although I may be lacking imagination here).
An obvious caveat to bigbrain is that it is a single brain and we know there are sometimes substantial individual variations in e.g. areal definition. This is only slightly touched upon in the discussion. Might be worth commenting on this more. As I see it, there are multiple considerations. For example (i) Surface-to-Surface registration in the presence of morphological idiosyncracies: what parts of the brain can we "trust" and what parts are uncertain? (ii) MRI parcellations mapped onto BigBrain will vary in how accurately they may reflect the BigBrain areal boundaries: if histo boundaries do not correspond with MRI-derived ones, is that because BigBrain is slightly different or is it a genuine divergence between modalities? Of course addressing these questions is out of scope of this manuscript, but some discussion could be useful; I also think this toolbox may be useful for addressing this very concerns!
-

Reviewer #2 (Public Review):
This is a nice paper presenting a review of recent developments and research resulting from BigBrain and a tutorial guiding use of the BigBrainWarp toolbox. This toolbox supports registration to, and from, standard MRI volumetric and surface templates, together with mapping derived features between spaces. Examples include projecting histological gradients estimated from BigBrain onto fsaverage (and the ICMB2009 atlas) and projecting Yeo functional parcels onto the BigBrain atlas.
The key strength of this paper is that it supports and expands on a comprehensive tutorial and docker support available from the website. The tutorials there go into even more detail (with accompanying bash scripts) of how to run the full pipelines detailed in the paper. The docker makes the tool very easy to install but I was also …
Reviewer #2 (Public Review):
This is a nice paper presenting a review of recent developments and research resulting from BigBrain and a tutorial guiding use of the BigBrainWarp toolbox. This toolbox supports registration to, and from, standard MRI volumetric and surface templates, together with mapping derived features between spaces. Examples include projecting histological gradients estimated from BigBrain onto fsaverage (and the ICMB2009 atlas) and projecting Yeo functional parcels onto the BigBrain atlas.
The key strength of this paper is that it supports and expands on a comprehensive tutorial and docker support available from the website. The tutorials there go into even more detail (with accompanying bash scripts) of how to run the full pipelines detailed in the paper. The docker makes the tool very easy to install but I was also able to install from source. The tutorials are diverse examples of broad possible applications; as such the combined resource has the potential to be highly impactful.
The minor weaknesses of the paper relate to its clarity and depth. Firstly, I found the motivations of the paper initially unclear from the abstract. I would recommend much more clearly stating that this is a review paper of recent research developments resulting from the BigBrain atlas, and a tutorial to accompany the bash scripts which apply the warps between spaces. The registration methodology is explained elsewhere.
I also found parts of the paper difficult to follow - as a methodologist without comprehensive neuroanatomical terminology, I would recommend the review of past work to be written in a more 'lay' way. In many cases, the figure captions also seemed insufficient at first. For example it was not immediately obvious to me what is meant by 'mesiotemporal confluence' and Fig 1G is not referenced specifically in the text. In Fig 3C it is not immediately clear from the text of the caption that the cortical image is representing the correlation from the plots - specifically since functional connectivity is itself estimated through correlation.
My minor concern is over the lack of details in relation to the registration pipelines. I understand these are either covered in previous papers or are probably destined for bespoke publications (in the case of the surface registration approach) but these details are important for readers to understand the constraints and limitations of the software. At this time, the details for the surface registration only relate to an OHBM poster and not a publication, which I was unable to find online until I went through the tutorial on the BigBrain website. In general I think a paper should have enough information on key techniques to stand alone without having to reference other publications, so, in my opinion, a high level review of these pipelines should be added here.
There isn't enough details on the registration. For the surface, what features were used to drive alignment, how was it parameterised (in particular the regularisation - strain, pairwise or areal), how was it pre-processed prior to running MSM - all these details seem to be in the excellent poster. I appreciate that work deserves a stand alone publication but some details are required here for users to understand the challenges, constraints and limitations of the alignment. Similar high level details should be given for the registration work.
I would also recommend more guidance in terms of limitations relating to inter-subject variation. My interpretation of the results of tutorial 3, is that topographic variation of the cortex could easily be driving the greater variation of the frontal parietal networks. Either that, or the Yeo parcel has insufficient granularity; however, in that case any attempt to go to finer MRI driven parcellations - for example to the HCP parcellation, would create its own problems due to subject specific variability.
-

Reviewer #3 (Public Review):
The authors make a point for the importance of considering high-resolution, cell-scale, histological knowledge for the analysis and interpretation of low-resolution MRI data. The manuscript describes the aims and relevance of the BigBrain project. The BigBrain is the whole brain of a single individual, sliced at 20µ and scanned at 1µ resolution. During the last years, a sustained work by the BigBrain team has led to the creation of a precise cell-scale, 3D reconstruction of this brain, together with manual and automatic segmentations of different structures.
The manuscript introduces a new tool - BigBrainWarp - which consolidates several of the tools used to analyse BigBrain into a single, easy to use and well documented tool. This tool should make it easy for any researcher to use the wealth of information …Reviewer #3 (Public Review):
The authors make a point for the importance of considering high-resolution, cell-scale, histological knowledge for the analysis and interpretation of low-resolution MRI data. The manuscript describes the aims and relevance of the BigBrain project. The BigBrain is the whole brain of a single individual, sliced at 20µ and scanned at 1µ resolution. During the last years, a sustained work by the BigBrain team has led to the creation of a precise cell-scale, 3D reconstruction of this brain, together with manual and automatic segmentations of different structures.
The manuscript introduces a new tool - BigBrainWarp - which consolidates several of the tools used to analyse BigBrain into a single, easy to use and well documented tool. This tool should make it easy for any researcher to use the wealth of information available in the BigBrain for the annotation of their own neuroimaging data.
The authors provide three examples of utilisation of BigBrainWarp, and show the way in which this can provide additional insight for analysing and understanding neuroimaging data.
The BigBrainWarp tool should have an important impact for neuroimaging research, helping bridge the multi-scale resolution gap, and providing a way for neuroimaging researchers to include cell-scale phenomena in their study of brain data.
All data and code are available open source, open access.Main concern:
One of the longstanding debates in the neuroimaging community concerns the relationship between brain geometry (in particular gyro/sulcal anatomy) and the cytoarchitectonic, connective and functional organisation of the brain. There are various examples of correspondance, but also many analyses showing its absence, particularly in associative cortex (for example, Fischl et al (2008) by some of the co-authors of the present manuscript). The manuscript emphasises the accuracy of their transformations to the different atlas spaces, which may give some readers a false impression. True: towards the end of the manuscript the authors briefly indicate the difficulty of having a single brain as source of histological data. I think, however, that the manuscript would benefit from making this point more clearly, providing the future users of BigBrainWarp with some conceptual elements and references that may help them properly apprise their results. In particular, it would be helpful to briefly describe which aspects of brain organisation where used to lead the deformation to the different templates, if they were only based on external anatomy, or if they took into account some other aspects such as myelination, thickness, ...
Minor:
In the abstract and later in p9 the authors talk about "state-of-the-art" non-linear deformation matrices. This may be confusing for some readers. To me, in brain imaging a matrix is most often a 4x4 affine matrix describing a linear transformation. However, the authors seem to be describing a more complex, non-linear deformation field. Whereas building a deformation matrix (4x4 affine) is not a big challenge, I agree that more sophisticated tools should provide more sophisticated deformation fields. The authors may consider using "deformation field" instead of "deformation matrix", but I leave that to their judgment.
In the results section, p11, the authors highlight the challenge of segmenting thalamic nuclei or different hippocampal regions, and suggest that this should be simplified by the use of the histological BigBrain data. However, the atlases currently provided in the OSF project do not include these more refined parcellation: there's one single "Thalamus" label, and one single "Hippocampus" label (not really single: left and right). This could be explicitly stated to prevent readers from having too high expectations (although I am certain that those finer parcellations should come in the very close future).
-