Protein allocation and utilization in the versatile chemolithoautotroph Cupriavidus necator
Curation statements for this article:-
Curated by eLife
Evaluation Summary:
This work combines elegant experimental approaches with modelling predictions to study of metabolic adaptations in the bacterium Cupriavidus necator, a microorganism of interest given its metabolic versatility and potential industrial applications. This manuscript will be interesting for microbiologists and systems biologists who want to understand how protein production and economy and enzyme utilization differs in a versatile microorganism in different conditions.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #1 and Reviewer #2 agreed to share their names with the authors.)
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Bacteria must balance the different needs for substrate assimilation, growth functions, and resilience in order to thrive in their environment. Of all cellular macromolecules, the bacterial proteome is by far the most important resource and its size is limited. Here, we investigated how the highly versatile 'knallgas' bacterium Cupriavidus necator reallocates protein resources when grown on different limiting substrates and with different growth rates. We determined protein quantity by mass spectrometry and estimated enzyme utilization by resource balance analysis modeling. We found that C. necator invests a large fraction of its proteome in functions that are hardly utilized. Of the enzymes that are utilized, many are present in excess abundance. One prominent example is the strong expression of CBB cycle genes such as Rubisco during growth on fructose. Modeling and mutant competition experiments suggest that CO 2 -reassimilation through Rubisco does not provide a fitness benefit for heterotrophic growth, but is rather an investment in readiness for autotrophy.
Article activity feed
-

-

Author Response:
Reviewer #2 (Public Review):
In this work, authors investigated the versatility of the beta-proteobacterium Cupriavidus necator from the proteome perspective. For this purpose, they cultivated the microorganism in a chemostat using different limiting substrates (fructose, fructose with limited ammonia, formate and succinate) and under different dilution rates. Integration of experimental proteomic data with a resource balance analysis model allowed to understand the relation between enzyme abundances and metabolic fluxes in the central metabolism. Moreover, the use of a transposon mutant library and competition experiments, could add insights regarding the essentiality of the genes studied. This shed light on the (under)utilization of metabolic enzymes, including some interpretations and speculations regarding C. …
Author Response:
Reviewer #2 (Public Review):
In this work, authors investigated the versatility of the beta-proteobacterium Cupriavidus necator from the proteome perspective. For this purpose, they cultivated the microorganism in a chemostat using different limiting substrates (fructose, fructose with limited ammonia, formate and succinate) and under different dilution rates. Integration of experimental proteomic data with a resource balance analysis model allowed to understand the relation between enzyme abundances and metabolic fluxes in the central metabolism. Moreover, the use of a transposon mutant library and competition experiments, could add insights regarding the essentiality of the genes studied. This shed light on the (under)utilization of metabolic enzymes, including some interpretations and speculations regarding C. necator's physiological readiness to changes in nutrients within its environmental niche. However, several parts of C. necator metabolism are not yet well analyzed (PHB biosynthesis and photorespiration) and some conclusions are not well reported.
Strengths:
- The manuscript is well written, easily understandable also for (pure) experimentalists, and adds a novel layer of comprehension in the physiology and metabolism of this biotechnologically relevant microorganism. Therefore, it is likely to raise attention and be well-cited among the metabolic engineering community of this organisms.
- More generally, the scope of the study is broad enough to potentially attract experts in the wider-field of autotrophic/mixotrophic metabolism, especially regarding the metabolic difference in the transition from heterotrophic to autotrophic growth modes and vice versa.
- Findings from different experimental techniques (chemostat cultivation, proteomics, modelling, mutant libraries) complement each other and increase the level of understanding. Consistency of the results from these different angles increases the roundness of the study.
Weaknesses:
- A main conclusion of this paper is that it concludes that the CCB cycle operation in heterotrophic conditions (fructose and succinate) is not useful for the biomass growth. However, Shimizu et al., 2015 claim that the CBB cycle has a benefit for at least PHB production is increased, in the presence of the CCB cycle (as demonstrated by a decrease in PHB production when Rubisco or cbbR are knocked out). In this work the authors do not analyze PHB production, but they do analyze fitness in mutant libraries. They claim not see this benefit in this study, however in their data (Figure 5 F) also small fitness drops are seen for cbbR mutants on fructose, as well as on succinate. So I think the authors have to revisit this conclusion. The type of modelling they use (RBA/FBA) may not explain such re-assimilation as 'a theoretically efficient' route, as this type of modelling assumes ' stochiometric' metabolic efficiency with setting a maximum growth objective, which is not what seems to happen in reality fully.
We agree that a minor decrease in fitness is visible for cbbR transposon mutants in heterotrophic conditions (Figure 5F). However, we have noticed that small changes in fitness can occur -particularly at a late stage of cultivation- as an artifact of the sequencing method (fast growing mutants displacing slow-growing ones). A replication of the experiment with pulsed instead of continuous feed showed a slightly increased instead of decreased fitness on succinate for cbbR (Figure 5-figure supplement 1). We therefore conclude that the resolution of the transposon library experiments is not sufficient to decide if the cbbR KO mutant conveys a small fitness benefit or loss. As the reviewer correctly points out, Shimizu et al. do not show a general fitness benefit but only increased PHB yield from CO2-refixation. We have rewritten our conclusions to account for the fact that our results do not contradict the findings from Shimizu et al., but that both increased PHB production and slightly decreased fitness (= growth rate) is possible at the same time. We also toned down our conclusions such that the question of a potential small fitness burden/benefit of the CBB cycle in heterotrophic conditions remains open.
- The authors focus a lot on readiness as a rational, but actually cannot really prove readiness as an explanation of the expression of 'unutilized' proteome, in the manuscript they also mention that it maybe a non-optimized, recent evolutionary trait, especially for the Calvin Cycle (especially because of the observed responsiveness to PEP of the cbbR regulator). The authors should discuss and not present as if readiness is the proven hypothesis. It would be interesting (and challenging) if the authors can come up with some further suggestions how to research and potentially proof readiness or ' evolutionary inefficiency'.
We rephrased the respective sections to highlight readiness as one potential explanation among others. We added a suggestion for an experimental strategy to test this hypothesis (laboratory evolution of lean-proteome strains).
- C. necator is well-known for the production of the storage polymer polyhydroxybutyrate (PHB) under nutrient-limited conditions, such as nitrogen of phosphate starvation. Even though the authors looked at such a nitrogen-limited condition ("ammonia") they do not report on the enzymes involved in this metabolism (phABC), which can be typically very abundant under these conditions. This should be discussed and ideally also analyzed. The formation of storage polymers is hard to incorporate in the flux balance analyze with growth as objective, however in real life C. necator can incorporate over 50% of carbon in PHB rather than biomass, so I suggest the authors discuss this and ideally develop a framework to analyze this, specifically for the ammonia-limited condition
As mentioned above to Reviewer 1, we have now performed nitrogen-limited chemostat cultivations in order to disentangle the formation of biomass and PHB. We have updated our model by incorporating separate fluxes 1) to biomass, and 2) to PHB according to the experimental results. We have also analyzed the enzyme abundance and utilization for phaA (in the model reaction ACACT1r), phaB (AACOAR) and phaC (PHAS). The first two enzymes showed high abundance that increased with degree of limitation for all substrates. PHAS showed a different pattern with much lower, constant expression. All enzymes were expressed regardless of N- or C-limitation, but the model did only show utilization during N-limitation where PHB production was enforced. These results were summarized in the new Figure 3-figure supplement 2.
- The authors extensively discuss the CCB cycle and its proteome abundance. However during autotrophic growth also typically photorespiration/phosphoglycolate salvage pathways are required to deal with the oxygenase side-activity of Rubisco. The authors have not discuss the abundance of the enzymes involved in that key process. Recently, a publication in PNAS on C. necator showed by transcriptomics and knockout that the glycerate pathway on hydrogen and low CO2 is highly abundant (10.1073/pnas.2012288117). Would be good to include these enzymes and the oxygenase side-activity in the modelling, proteome analysis and fitness analysis. An issue with the growth on formate is that the real CO2 concentration in the cells cannot be determined well, but not feeding additional CO2, likely results in substantial oxygenase activity
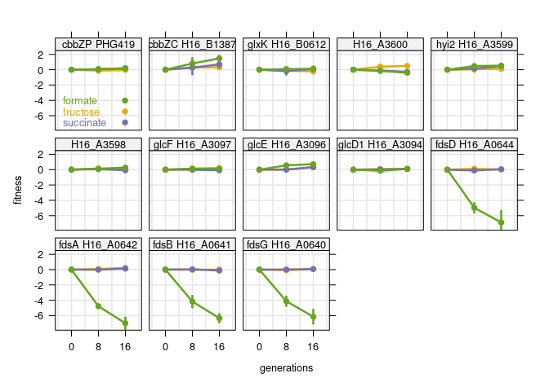
C. necator has several pathways for 2-phosphoglycolate (2-PGly) salvage, as the reviewer points out. The key enzymes for the universal 'upper part' of 2-PGly salvage, 2-PGly-phosphatase (cbbZ2, cbbZP) and glycolate dehydrogenase GDH (GlcDEF), were all quantified in our proteomics experiments. The cbbZ isoenzymes showed identical expression compared to the other cbb enzymes: highest on formate, lowest on succinate (Figure 1-figure supplement 2D). The GDH subunits encoded by GlcDEF showed no significant trend between growth rates or substrates, and were more than 10-fold lower abundant than 2-PGly-phosphatase. This is in line with the findings from Claassens et al., PNAS, 2020, that showed only a 2.5-fold upregulation of GDH transcripts in a low versus high CO2 comparison (changes on protein level are often less extreme than transcript). The same study demonstrated that the glycerate pathway is the dominant route for 2-PGly salvage and found four enzymes extremely upregulated in low CO2: glyoxylate carboligase GLXCL (H16_A3598), hydroxypyruvate isomerase HPYRI (H16_A3599), tartronate semialdehyde reductase TRSARr (H16_A3600), and glycerate kinase GLYCK (H16_B0612). Here, these enzymes showed only slightly higher abundance on formate compared to the other conditions we tested (~2-fold). The increase was much lower than what the transcriptional upregulation in Classens et al. would suggest; It is therefore difficult to say if 2-PGly salvage plays a role during formatotrophic growth. Moreover, we also investigated conditional essentiality and found that none of the 2-PGly salvage mutants showed impaired growth on formate (see Figure R1 below).
Unfortunately there is -to our knowledge- no data available on the rate of Rubisco's oxygenation reaction during formatotrophic growth, and our bioreactor setup does not support measurement of pCO2. It is known though that only 25% of the CO2 from formic acid oxidation is consumed for biomass (Grunwald et al., Microb Biotech, 2015, http://dx.doi.org/10.1111/1751-7915.12149), effectively creating an excess intracellular CO2 supply. Further, the substrate specificity of the C. necator Rubisco for CO2 over O2 is very high, about twice that of cyanobacteria (Horken & Tabita, Arch Biochem Biophys, 1999, https://pubmed.ncbi.nlm.nih.gov/9882445/). This indirect evidence suggests that flux through this pathway is most likely marginal. We therefore decided to omit it from model simulations. We have added a paragraph summarizing our findings regarding phosphoglycolate salvage to the results section.
Figure R1: Fitness of 2-phosphoglycolate salvage mutants during growth on three different carbon sources, fructose, formate, and succinate. Four genes essential for growth on formate were included for comparison (soluble formate dehydrogenase fdsABDG). Fitness scores are mean and standard deviation of four biological replicates.
-

Evaluation Summary:
This work combines elegant experimental approaches with modelling predictions to study of metabolic adaptations in the bacterium Cupriavidus necator, a microorganism of interest given its metabolic versatility and potential industrial applications. This manuscript will be interesting for microbiologists and systems biologists who want to understand how protein production and economy and enzyme utilization differs in a versatile microorganism in different conditions.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #1 and Reviewer #2 agreed to share their names with the authors.)
-

Reviewer #1 (Public Review):
The study describes in detail protein abundance and utilization in C. necator. The authors show highly interesting data and draw conclusions about the strategy of C. necator to be ready for environmental changes. The authors combine experimental data and models to predict environmental adpatation of C. necator and the enzyme utilisation. Especially the data regarding the adapation of the CBB in the new chromosome and plasmid are of special interest.
The data are presented in a good fashion and the methods are described sufficient.
The hypothesis of enzyme utilization was modeled, tested and analyzed and showed a clear different strategy compared to E. coli. The barcode mutant library is a excellent tool to test enzyme essentiality. Interesting is the high protein abundance of unutilized enzymes.
The authors …Reviewer #1 (Public Review):
The study describes in detail protein abundance and utilization in C. necator. The authors show highly interesting data and draw conclusions about the strategy of C. necator to be ready for environmental changes. The authors combine experimental data and models to predict environmental adpatation of C. necator and the enzyme utilisation. Especially the data regarding the adapation of the CBB in the new chromosome and plasmid are of special interest.
The data are presented in a good fashion and the methods are described sufficient.
The hypothesis of enzyme utilization was modeled, tested and analyzed and showed a clear different strategy compared to E. coli. The barcode mutant library is a excellent tool to test enzyme essentiality. Interesting is the high protein abundance of unutilized enzymes.
The authors also sufficiently describe the limitations of the study, especially the unanotated proteins and proteins not involved in pathways (sensing, motion etc.) -

Reviewer #2 (Public Review):
In this work, authors investigated the versatility of the beta-proteobacterium Cupriavidus necator from the proteome perspective. For this purpose, they cultivated the microorganism in a chemostat using different limiting substrates (fructose, fructose with limited ammonia, formate and succinate) and under different dilution rates. Integration of experimental proteomic data with a resource balance analysis model allowed to understand the relation between enzyme abundances and metabolic fluxes in the central metabolism. Moreover, the use of a transposon mutant library and competition experiments, could add insights regarding the essentiality of the genes studied. This shed light on the (under)utilization of metabolic enzymes, including some interpretations and speculations regarding C. necator's physiological …
Reviewer #2 (Public Review):
In this work, authors investigated the versatility of the beta-proteobacterium Cupriavidus necator from the proteome perspective. For this purpose, they cultivated the microorganism in a chemostat using different limiting substrates (fructose, fructose with limited ammonia, formate and succinate) and under different dilution rates. Integration of experimental proteomic data with a resource balance analysis model allowed to understand the relation between enzyme abundances and metabolic fluxes in the central metabolism. Moreover, the use of a transposon mutant library and competition experiments, could add insights regarding the essentiality of the genes studied. This shed light on the (under)utilization of metabolic enzymes, including some interpretations and speculations regarding C. necator's physiological readiness to changes in nutrients within its environmental niche. However, several parts of C. necator metabolism are not yet well analyzed (PHB biosynthesis and photorespiration) and some conclusions are not well reported.
Strengths:
1. The manuscript is well written, easily understandable also for (pure) experimentalists, and adds a novel layer of comprehension in the physiology and metabolism of this biotechnologically relevant microorganism. Therefore, it is likely to raise attention and be well-cited among the metabolic engineering community of this organisms.
2. More generally, the scope of the study is broad enough to potentially attract experts in the wider-field of autotrophic/mixotrophic metabolism, especially regarding the metabolic difference in the transition from heterotrophic to autotrophic growth modes and vice versa.
3. Findings from different experimental techniques (chemostat cultivation, proteomics, modelling, mutant libraries) complement each other and increase the level of understanding. Consistency of the results from these different angles increases the roundness of the study.
Weaknesses:
1. A main conclusion of this paper is that it concludes that the CCB cycle operation in heterotrophic conditions (fructose and succinate) is not useful for the biomass growth. However, Shimizu et al., 2015 claim that the CBB cycle has a benefit for at least PHB production is increased, in the presence of the CCB cycle (as demonstrated by a decrease in PHB production when Rubisco or cbbR are knocked out). In this work the authors do not analyze PHB production, but they do analyze fitness in mutant libraries. They claim not see this benefit in this study, however in their data (Figure 5 F) also small fitness drops are seen for cbbR mutants on fructose, as well as on succinate. So I think the authors have to revisit this conclusion. The type of modelling they use (RBA/FBA) may not explain such re-assimilation as 'a theoretically efficient' route, as this type of modelling assumes ' stochiometric' metabolic efficiency with setting a maximum growth objective, which is not what seems to happen in reality fully.
2. The authors focus a lot on readiness as a rational, but actually cannot really prove readiness as an explanation of the expression of 'unutilized' proteome, in the manuscript they also mention that it maybe a non-optimized, recent evolutionary trait, especially for the Calvin Cycle (especially because of the observed responsiveness to PEP of the cbbR regulator). The authors should discuss and not present as if readiness is the proven hypothesis. It would be interesting (and challenging) if the authors can come up with some further suggestions how to research and potentially proof readiness or ' evolutionary inefficiency'.
3. C. necator is well-known for the production of the storage polymer polyhydroxybutyrate (PHB) under nutrient-limited conditions, such as nitrogen of phosphate starvation. Even though the authors looked at such a nitrogen-limited condition ("ammonia") they do not report on the enzymes involved in this metabolism (phABC), which can be typically very abundant under these conditions. This should be discussed and ideally also analyzed. The formation of storage polymers is hard to incorporate in the flux balance analyze with growth as objective, however in real life C. necator can incorporate over 50% of carbon in PHB rather than biomass, so I suggest the authors discuss this and ideally develop a framework to analyze this, specifically for the ammonia-limited condition
4. The authors extensively discuss the CCB cycle and its proteome abundance. However during autotrophic growth also typically photorespiration/phosphoglycolate salvage pathways are required to deal with the oxygenase side-activity of Rubisco. The authors have not discuss the abundance of the enzymes involved in that key process. Recently, a publication in PNAS on C. necator showed by transcriptomics and knockout that the glycerate pathway on hydrogen and low CO2 is highly abundant (10.1073/pnas.2012288117). Would be good to include these enzymes and the oxygenase side-activity in the modelling, proteome analysis and fitness analysis. An issue with the growth on formate is that the real CO2 concentration in the cells cannot be determined well, but not feeding additional CO2, likely results in substantial oxygenase activity.
-

Reviewer #3 (Public Review):
This work provides new knowledge regarding the versatile chemolithoautotrophic bacterium C. necator, such as presence of many under-utilized enzymes as well as excess amount of utilized enzymes, suggesting the highly robust properties of this bacterium against environmental perturbations. One weakness is thought to be lack of consideration into polyhydroxyalkanoate synthesis occurred in this bacterium under nitrogen limited condition.
-