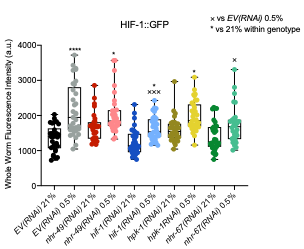Nuclear hormone receptor NHR-49 acts in parallel with HIF-1 to promote hypoxia adaptation in Caenorhabditis elegans
Curation statements for this article:-
Curated by eLife
Evaluation Summary:
This study brings new insight into how organisms maintain homeostasis under stress conditions and has implications for our understanding both development and disease. The study provides evidence that NHR-49 protects animals from hypoxia by activating autophagy, and that it acts independently of the well-described canonical HIF-1 hypoxia response. The experiments are well done, and the conclusions from the results are largely appropriate. The impact of this study will be highest in the specific field of hypoxia, with more moderate impact for wider audiences interested in understanding of how biological maintain homeostasis under stress.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. The reviewers remained anonymous to the authors.)
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
The response to insufficient oxygen (hypoxia) is orchestrated by the conserved hypoxia-inducible factor (HIF). However, HIF-independent hypoxia response pathways exist that act in parallel with HIF to mediate the physiological hypoxia response. Here, we describe a hypoxia response pathway controlled by Caenorhabditis elegans nuclear hormone receptor NHR-49, an orthologue of mammalian peroxisome proliferator-activated receptor alpha (PPARα). We show that nhr-49 is required for animal survival in hypoxia and is synthetic lethal with hif-1 in this context, demonstrating that these factors act in parallel. RNA-seq analysis shows that in hypoxia nhr-49 regulates a set of genes that are hif-1- independent, including autophagy genes that promote hypoxia survival. We further show that nuclear hormone receptor nhr-67 is a negative regulator and homeodomain-interacting protein kinase hpk-1 is a positive regulator of the NHR-49 pathway. Together, our experiments define a new, essential hypoxia response pathway that acts in parallel with the well-known HIF-mediated hypoxia response.
Article activity feed
-

-

Author Response:
Reviewer #1 (Public Review):
Summary
The authors have discovered and characterized a novel genetic pathway responsive to hypoxia, which acts in parallel to the canonical response through activation of Hypoxia-Inducible Factor (HIF). Specifically, the authors discovered that the Caenorhabditis elegans nuclear hormone receptor NHR-49, ortholog to mammalian PPAR-alpha, is essential for survival under hypoxic conditions and regulates target gene expression that is hif-1-independent; identifying an essential role of autophagy. Further the authors discover both positive and negative regulators of NHR-49 and a putative feedback loop.
Overall analysis
The genetic analysis conducted by the authors is outstanding. However, the study is lacking in a few key areas and the authors may have over-interpreted results in a few …
Author Response:
Reviewer #1 (Public Review):
Summary
The authors have discovered and characterized a novel genetic pathway responsive to hypoxia, which acts in parallel to the canonical response through activation of Hypoxia-Inducible Factor (HIF). Specifically, the authors discovered that the Caenorhabditis elegans nuclear hormone receptor NHR-49, ortholog to mammalian PPAR-alpha, is essential for survival under hypoxic conditions and regulates target gene expression that is hif-1-independent; identifying an essential role of autophagy. Further the authors discover both positive and negative regulators of NHR-49 and a putative feedback loop.
Overall analysis
The genetic analysis conducted by the authors is outstanding. However, the study is lacking in a few key areas and the authors may have over-interpreted results in a few places, which diminishes my overall enthusiasm. These concerns are addressable and doing so would greatly strengthen the manuscript. I highlight individual major concerns below, and save minor concerns and specific suggestions for private recommendations for the authors.
Major concerns
1 The authors have provided strong genetic evidence for a parallel mechanism to canonical HIF-1 activity in response to hypoxia. The authors should more rigorously test whether there is evidence for cross-talk between the two mechanisms. In the discussion the authors' highlight findings in mammals that support this possibility. For example, does loss of one lead to hyperactivation of the other in an attempt to compensate for hypoxia?
We thank the reviewer for suggesting these interesting experiments to examine cross-talk!
Specific examples:
• In regards to lines 425-426, does loss of hpk-1 stabilize HIF-1 (or does hpk-1(oe) repress hif-1)?
We attempted to study HPK-1–HIF-1 cross talk via GFP imaging of the UL1447 HIF-1::GFP strain after hpk-1 RNAi (Figure R4, below). However, although we did observe an increase in GFP levels in hypoxia (vs. normoxia), we did not observe nuclear localization, possibly due to the rapid degradation of HIF-1 in normoxia, which occurs inevitably during our experimental procedure. We therefore opted not to include these data in the manuscript.
Figure R4: Regulation of HIF-1::GFP. Quantification of GFP levels in UL1447 (unc-119(ed3) III; leEx1447 [hif-1::GFP + unc-119(+)) adult animals expressing HIF-1::GFP. Animals were fed EV RNAi or nhr-49, hif-1, hpk-1, or nhr-67 RNAi as indicated and exposed to 4 hr of 0.5% O2 without recovery (three repeats totalling >30 individual animals per strain). X/, XXX,**** p <0.05, 0.001, 0.0001 (two-way ANOVA corrected for multiple comparisons using the Tukey method).
• Does loss of hif-1 or nhr-49 alter the expression, stability, or activity of the other (either under normoxic or hypoxic conditions)?
We appreciate the reviewer’s interest in examining the interaction between nhr-49 and hif-1. To address this, we generated an NHR-49::GFP;hif-1(-) strain and analysed it by imaging after exposure to normoxia or hypoxia. Although loss of hif-1 does result in a slight whole-body up-regulation of NHR-49::GFP, this increase was not significant (new Figure 2—figure supplement 1C, D). Higher magnification images did not show a tissue-specific effect in NHR-49::GFP increase in the hif-1(-) background either (new Figure 2D, Figure 2—figure supplement 1E, F). For reasons mentioned above, the HIF-1::GFP;nhr-49(RNAi) experiment was inconclusive.
• Can overexpression of either hif-1 or nhr-49 rescue the developmental defects caused by loss of the other (i.e. overexpress hif-1 in nhr-49 mutant animals, and vice versa).
With the new NHR-49::GFP;hif-1(-) strain, we were able to study compensatory effects of overexpressing NHR-49 in hif-1 mutants by performing embryo hypoxia survival experiments (new Figure 2E). Excitingly, while NHR-49 overexpression does not provide enhanced hypoxia survival at baseline (vs. non-GFP siblings), NHR-49 overexpression rescued the deficiency of hif-1 mutants. This suggests that nhr-49 can partially compensate for loss of the hif-1 pathway. Testing whether HIF-1::GFP overexpression rescues nhr-49 loss requires non-GFP sibling controls. Although the UL1447 strain expresses HIF-1::GFP from an extrachromosomal array, in our hands, we never observed non-GFP worms (i.e. 100% HIF-1::GFP offspring), and therefore were unable to test whether HIF-1 overexpression compensates for nhr-49 loss.
• Does NHR-67 negatively regulate hif-1 (specificity to NHR-49)?
As noted above, we were unfortunately unable to conclusively assessed HIF-1::GFP levels, likely due to rapid degradation during the normoxia that occurs during animal harvest.
2 The role of autophagy in hypoxia should be explored in greater detail. While the evidence presented by the authors clearly demonstrates autophagy is essential for hypoxic survival, autophagy is an important component of many biological processes. Thus, it's critical to distinguish whether autophagy is merely required (perhaps for very indirect reasons) or whether autophagy is a part of an adaptive response to hypoxia. The authors (Miller lab) previously failed to find a role for autophagy in hypoxia (Fawcett et al. 2015 Aging Cell), which should be addressed. Has autophagy been previously linked to hypoxia in C. elegans? The novelty of this discovery should be discussed in greater detail.
We appreciate that the link of autophagy to hypoxia survival needed to be examined further. We now provide substantial new evidence showing that not only are autophagy genes and autophagosome formation induced in hypoxia, but also that mutations in autophagy genes result in hypoxia sensitivity. In our opinion, this strongly supports a key role for autophagy in hypoxia adaptation.
We note that the study by Fawcett et al., 2015 studied only two genes in hypoxia, bec-1 and unc-51, none of which were found to be regulated by hypoxia in our RNA-seq analysis. Another study from the Miller lab found that an 18-hour anoxia exposure of L2/L3 stage C. elegans results in a significant induction of autophagy in the intestine (Chapin et al., 2015; Fig 4B, C). Although the conditions in this study are different than in ours (anoxia vs. hypoxia, exposure time, animal developmental stage), this study, like ours, thus finds that that low oxygen availability induces autophagy. Besides the Miller lab, there are several other publications that show an important role for autophagy in hypoxia adaptation across species (Samokhvalov et al., 2008; Zhang et al., 2008). Especially relevant to our manuscript is a recent paper published while we were revising our manuscript, which shows that autophagy gene induction is HIF-1 independent in Drosophila melanogaster (Valko et al., 2021). This agrees well with our exciting new discoveries. We have revised the text to better discuss this context.
3 The authors have possibly over-interpreted their results in Figure 4B and the possibility that NHR-49 acts cell non-autonomously. The authors speculate that tissue specific genetic rescue by NHR-49 over-expression could indicate the existence of a signaling molecule (line 499). Ectopic over-expression of a transcription factor within one tissue is always tricky to interpret, as it may not be physiologically relevant, which I fear may be the case as rescue is achieved when NHR-49 is over-expressed within any tissue (i.e. there is no specificity). An alternative explanation, which is a more indirect model, is that NHR-49 over-expression shifts metabolism within a tissue to generate metabolites that are released throughout the organism to sustain it during hypoxia.
We thank the reviewer for this excellent point, and agree that indirect action of NHR-49 remains a possibility. We have added discussion to this point in the revised manuscript.
4 As an extension of MC#3, the authors demonstrate that NHR-49 is induced throughout the animal after hypoxia (Figure 5A). Presumably sites of NHR-49 induction (tissues) equates to the sites where nhr-49 is necessary. However, the images within 5A cannot be resolved to identify individual tissues, higher resolution images are necessary and quantification of GFP expression within individual tissues could lend biological insight.
We now provide higher resolution images of NHR-49::GFP in Figure 2D, Figure 2—figure supplement 1E, F.
5 The gene expression analysis is lacking details. For example, the RNA-seq data shown in Figure 3A&B is confusing. The numbers in the text do not match the figure and it is unclear whether the intersection in the Venn Diagram represent inverse relationships (i.e. the proportion of genes that are upregulated in wild-type that are either hif-1 or nhr-49 dependent). Greater detail and explanation is needed, as presented little biological insight can be discerned from the Figure 3A&B. Next, qRT-PCR validation of autophagy gene expression found in Figure 3C should be provided with that result. Lastly, are there existing datasets for changes in gene expression of C. elegans exposed to hypoxia? If so, how do the datasets compare?
We apologize for the confusion and have revised our text describing the RNA-seq analysis as well as the Figure legend. We also provide validation of the RNA-seq data with GFP-reporters and have compared our dataset to a previous study on hypoxia dependent gene regulation in C. elegans.
6 The authors identify a putative negative feedback loop between NHR-67 and NHR-49, and suggest this regulation is at the protein level (Figure 5F,G) based on a translational reporter and not transcriptional regulation based on qRT-PCR results and similar results previously found with hpk-1 (Figures S5A, 7a, and a previous study). However, the authors should more rigorously rule out dynamic changes in expression between tissues that cannot be ascertained by qRT-PCR (i.e. test whether nhr-49p::GFP expression is altered after nhr-67(RNAi) +/- hypoxia.
We agree and have more rigorously studied this interaction.
-

Evaluation Summary:
This study brings new insight into how organisms maintain homeostasis under stress conditions and has implications for our understanding both development and disease. The study provides evidence that NHR-49 protects animals from hypoxia by activating autophagy, and that it acts independently of the well-described canonical HIF-1 hypoxia response. The experiments are well done, and the conclusions from the results are largely appropriate. The impact of this study will be highest in the specific field of hypoxia, with more moderate impact for wider audiences interested in understanding of how biological maintain homeostasis under stress.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. …
Evaluation Summary:
This study brings new insight into how organisms maintain homeostasis under stress conditions and has implications for our understanding both development and disease. The study provides evidence that NHR-49 protects animals from hypoxia by activating autophagy, and that it acts independently of the well-described canonical HIF-1 hypoxia response. The experiments are well done, and the conclusions from the results are largely appropriate. The impact of this study will be highest in the specific field of hypoxia, with more moderate impact for wider audiences interested in understanding of how biological maintain homeostasis under stress.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. The reviewers remained anonymous to the authors.)
-

Reviewer #1 (Public Review):
Summary:
The authors have discovered and characterized a novel genetic pathway responsive to hypoxia, which acts in parallel to the canonical response through activation of Hypoxia-Inducible Factor (HIF). Specifically, the authors discovered that the Caenorhabditis elegans nuclear hormone receptor NHR-49, ortholog to mammalian PPAR-alpha, is essential for survival under hypoxic conditions and regulates target gene expression that is hif-1-independent; identifying an essential role of autophagy. Further the authors discover both positive and negative regulators of NHR-49 and a putative feedback loop.
Overall analysis:
The genetic analysis conducted by the authors is outstanding. However, the study is lacking in a few key areas and the authors may have over-interpreted results in a few places, which diminishes …
Reviewer #1 (Public Review):
Summary:
The authors have discovered and characterized a novel genetic pathway responsive to hypoxia, which acts in parallel to the canonical response through activation of Hypoxia-Inducible Factor (HIF). Specifically, the authors discovered that the Caenorhabditis elegans nuclear hormone receptor NHR-49, ortholog to mammalian PPAR-alpha, is essential for survival under hypoxic conditions and regulates target gene expression that is hif-1-independent; identifying an essential role of autophagy. Further the authors discover both positive and negative regulators of NHR-49 and a putative feedback loop.
Overall analysis:
The genetic analysis conducted by the authors is outstanding. However, the study is lacking in a few key areas and the authors may have over-interpreted results in a few places, which diminishes my overall enthusiasm. These concerns are addressable and doing so would greatly strengthen the manuscript. I highlight individual major concerns below, and save minor concerns and specific suggestions for private recommendations for the authors.
Major concerns:
1. The authors have provided strong genetic evidence for a parallel mechanism to canonical HIF-1 activity in response to hypoxia. The authors should more rigorously test whether there is evidence for cross-talk between the two mechanisms. In the discussion the authors' highlight findings in mammals that support this possibility. For example, does loss of one lead to hyperactivation of the other in an attempt to compensate for hypoxia? Specific examples:
• In regards to lines 425-426, does loss of hpk-1 stabilize HIF-1 (or does hpk-1(oe) repress hif-1)?
• Does loss of hif-1 or nhr-49 alter the expression, stability, or activity of the other (either under normoxic or hypoxic conditions)?
• Can overexpression of either hif-1 or nhr-49 rescue the developmental defects caused by loss of the other (i.e. overexpress hif-1 in nhr-49 mutant animals, and vice versa).
• Does NHR-67 negatively regulate hif-1 (specificity to NHR-49)?2. The role of autophagy in hypoxia should be explored in greater detail. While the evidence presented by the authors clearly demonstrates autophagy is essential for hypoxic survival, autophagy is an important component of many biological processes. Thus, it's critical to distinguish whether autophagy is merely required (perhaps for very indirect reasons) or whether autophagy is a part of an adaptive response to hypoxia. The authors (Miller lab) previously failed to find a role for autophagy in hypoxia (Fawcett et al. 2015 Aging Cell), which should be addressed. Has autophagy been previously linked to hypoxia in C. elegans? The novelty of this discovery should be discussed in greater detail.
3. The authors have possibly over-interpreted their results in Figure 4B and the possibility that NHR-49 acts cell non-autonomously. The authors speculate that tissue specific genetic rescue by NHR-49 over-expression could indicate the existence of a signaling molecule (line 499). Ectopic over-expression of a transcription factor within one tissue is always tricky to interpret, as it may not be physiologically relevant, which I fear may be the case as rescue is achieved when NHR-49 is over-expressed within any tissue (i.e. there is no specificity). An alternative explanation, which is a more indirect model, is that NHR-49 over-expression shifts metabolism within a tissue to generate metabolites that are released throughout the organism to sustain it during hypoxia.
4. As an extension of MC#3, the authors demonstrate that NHR-49 is induced throughout the animal after hypoxia (Figure 5A). Presumably sites of NHR-49 induction (tissues) equates to the sites where nhr-49 is necessary. However, the images within 5A cannot be resolved to identify individual tissues, higher resolution images are necessary and quantification of GFP expression within individual tissues could lend biological insight.
5. The gene expression analysis is lacking details. For example, the RNA-seq data shown in Figure 3A&B is confusing. The numbers in the text do not match the figure and it is unclear whether the intersection in the Venn Diagram represent inverse relationships (i.e. the proportion of genes that are upregulated in wild-type that are either hif-1 or nhr-49 dependent). Greater detail and explanation is needed, as presented little biological insight can be discerned from the Figure 3A&B. Next, qRT-PCR validation of autophagy gene expression found in Figure 3C should be provided with that result. Lastly, are there existing datasets for changes in gene expression of C. elegans exposed to hypoxia? If so, how do the datasets compare?
6. The authors identify a putative negative feedback loop between NHR-67 and NHR-49, and suggest this regulation is at the protein level (Figure 5F,G) based on a translational reporter and not transcriptional regulation based on qRT-PCR results and similar results previously found with hpk-1 (Figures S5A, 7a, and a previous study). However, the authors should more rigorously rule out dynamic changes in expression between tissues that cannot be ascertained by qRT-PCR (i.e. test whether nhr-49p::GFP expression is altered after nhr-67(RNAi) +/- hypoxia.
-

Reviewer #2 (Public Review):
The data provided in the manuscript is mostly of good quality and the interpretations are sound. However, since the central message of this paper is characterization of this HIF-1-independent hypoxia response pathway, some more mechanistic detail needs to be provided. How the kinase HPK-1 activates NHR-49 specifically in hypoxic conditions needs some further investigation using biochemical approaches. In addition, the reciprocal inhibitory relationship between nhr-49 and nhr-67 during hypoxia should be explored a bit further because both the NHRs seem to promote survival in hypoxic conditions, but it is not clear whether they do so via the same or parallel pathways.
-

Reviewer #3 (Public Review):
Here, the authors identify a HIF-independent pathway controlled by NHR-49 in the C. elegans system. Authors hypothesized that nhr-49 has a role in hypoxia and regulates fmo-2 and genes involved with autophagy. Genetic analysis indicates that nhr-49 is required for hypoxia survival. Transcriptomic studies show that NHR-49 regulates a set of genes in hypoxia, that are HIF-1 independent. Authors provide evidence that NHR-67 is a negative regulator and HPK-1 is a positive regulator of the NHR-49 pathway. Authors found that autophagy gene dysfunction compromized hypoxia survival. It is unclear if the NHR-67 and HPK-1 regulators of NHR-49 impact the genes involved with autophagy as most transcriptional readout assays were done with the fmo-2 and acs-2 reporters. However, authors convincingly show that NHR-49 is …
Reviewer #3 (Public Review):
Here, the authors identify a HIF-independent pathway controlled by NHR-49 in the C. elegans system. Authors hypothesized that nhr-49 has a role in hypoxia and regulates fmo-2 and genes involved with autophagy. Genetic analysis indicates that nhr-49 is required for hypoxia survival. Transcriptomic studies show that NHR-49 regulates a set of genes in hypoxia, that are HIF-1 independent. Authors provide evidence that NHR-67 is a negative regulator and HPK-1 is a positive regulator of the NHR-49 pathway. Authors found that autophagy gene dysfunction compromized hypoxia survival. It is unclear if the NHR-67 and HPK-1 regulators of NHR-49 impact the genes involved with autophagy as most transcriptional readout assays were done with the fmo-2 and acs-2 reporters. However, authors convincingly show that NHR-49 is important for hypoxia responses, and functions in a HIF-1 independent/parallel pathway. It will be of interest to further investigate the role of NHR-49 and autophagy regulation in hypoxia survival.
-