Differences in the gut microbiomes of distinct ethnicities within the same geographic area are linked to host metabolic health
Curation statements for this article:-
Curated by eLife
Evaluation Summary:
This study aims to measure interpersonal and interethnic variation in human microbiomes and metabolomes in the San Franciso, CA, area. The strength of the study is in the level of robust analysis of the microbiota. It is interesting that diet is not one of the apparent associations in this study, yet the relationship of microbiota diversity to body habitus is strong in Caucasian subjects. Overall, the key results of this work nicely confirm that there are dissimilarities in gut microbiomes related to differences in ethnicity of subjects.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #3 agreed to share their name with the authors.)
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Background
The human gut microbiota exhibits marked variation around the world, which has been attributed to dietary intake and other environmental factors. However, the degree to which ethnicity-associated differences in gut microbial community structure and function are maintained following immigration or in the context of metabolic disease is poorly understood.
Results
We conducted a multi-omic study of 46 lean and obese East Asian and White participants living in the San Francisco Bay Area. 16S rRNA gene sequencing revealed significant differences between ethnic groups in bacterial richness and community structure. White individuals were enriched for the mucin-degrading Akkermansia muciniphila. East Asian participants had increased levels of multiple bacterial phyla, fermentative pathways detected by metagenomics, and the short-chain fatty acid end products acetate, propionate, and isobutyrate. Differences in the gut microbiota between the East Asian and White groups could not be explained by reported dietary intake, were more pronounced in lean individuals, and were associated with current geographical location. Microbiome transplantations into germ-free mice confirmed that the differences in the gut microbiota of the East Asian and White individuals we analyzed are independent of diet and that they differentially impact host body weight and adiposity in genetically identical mouse recipients.
Conclusions
The reported findings emphasize the utility of studying diverse ethnic groups within a defined geographical location and provide a starting point for dissecting the mechanisms contributing to the complex interactions between the gut microbiome and ethnicity-associated lifestyle, demographic, metabolic, and genetic factors.
Article activity feed
-

Author Response:
Reviewer #1 (Public Review):
The key question that the authors were addressing was how ethnicity differentially affects the microbiota of subjects living in a particular area (in this case East Asians and Caucasians living in San Francisco that have been enrolled in an 'Inflammation, Diabetes, Ethnicity and Obesity cohort - although inflammatory disease was apparently excluded in these subjects).
The existence of differences between different populations allows potential discrimination of the underlying factors - such as host genetics, diet, lifestyle, physiological parameters, body habitus or other environmental influences. In this case body habitus has been selected as a stratification factor between the two ethnicities. Immigration potentially allows distinction of environmental and host genetical influences.
T…
Author Response:
Reviewer #1 (Public Review):
The key question that the authors were addressing was how ethnicity differentially affects the microbiota of subjects living in a particular area (in this case East Asians and Caucasians living in San Francisco that have been enrolled in an 'Inflammation, Diabetes, Ethnicity and Obesity cohort - although inflammatory disease was apparently excluded in these subjects).
The existence of differences between different populations allows potential discrimination of the underlying factors - such as host genetics, diet, lifestyle, physiological parameters, body habitus or other environmental influences. In this case body habitus has been selected as a stratification factor between the two ethnicities. Immigration potentially allows distinction of environmental and host genetical influences.
The strength of the study is in the level of robust analysis of the microbiotas by a very experienced group of researchers, distinguishing the microbiota differences, especially in lean subject, with analysis of associations that may be driving the differences. It is interesting that diet is not one of the apparent associations in this study, yet the relationship of microbiota diversity to body habitus is strong in Caucasian subjects. These associations cannot easily be extrapolated to causation or mechanism - a fact well recognized in the paper - but remain important observations that rationalize in vivo modeling with experimental animals or in vitro analyses of microbial interactions between different taxa simulating the context of differences in the intestinal milieu. The paper includes work showing that differences of the microbiota can be recapitulated after transfer to germ-free mice, at least over the short term: this is important to provide tools to model the reasons for differences in consortial composition.
A very large amount of work required to assemble the samples and the clinical phenotypic metadata set making the data an important and definitive contribution for the subjects studied. Of course, this is one sample of extremely variable human conditions and lifestyles that will help build the overall picture of how differences in our genetics and environment shape our intestinal microbiota.
We appreciate the reviewers' positive summary of our manuscript and agree with the reviewer’s assessment of the need for both mechanistic follow-on studies and extensions to larger and more diverse cohorts.
Reviewer #2 (Public Review):
The study's primary aims are to test for the differences in the microbiome between self-identified East Asian and White subjects from the San Francisco area in the new IDEO cohort. The study builds on an growing literature which describes variations among ethnic groups. The major conclusion of "emphasize the utility of studying diverse ethnic groups" is not novel to the literature.
It was not our intention to imply that our study is novel in studying two distinct ethnic groups, but rather to emphasize that differences exist between ethnicities with regard to the gut microbiome and to provide a systematic analysis of this including gnotobiotic mouse models along a key health disparity in Asian Americans. We include references of prior examples of this work in our introduction (including several references in our introductory paragraph). We have modified our abstract to clarify this point further:
“Taken together, our findings add to the growing body of literature describing variation between ethnicities and provide a starting point for defining the mechanisms through which the microbiome may shape disparate health outcomes in East Asians.”
Overall, the strength of the results is that they confirm patterns from different cohorts/studies and demonstrate that ethnic-related differences are common. The results are subject to sample size concerns that may underpin some of the conflicting or lack of significant results. For instance, there is no overlap in highlighted species-level taxonomy differences between 16S and metagenomic analyses, which precludes a clear interpretation of the meaning of those differences and whether taxa should be highlighted in the abstract; there are low AUC values for the random forest modelling; and there is a lack of significance in correlations between BMI and East Asian subjects in F4a where there may be a correlation. While a minor point, it serves to highlight the sample sizes as the range of the variation in East Asian subjects is not as substantial as the White subjects because there are fewer East Asian data points above a 30 BMI (~N=5) relative to those of White subjects (~N=11).
We agree that our study was limited by sample size and that future studies increasing sample size would be valuable to assess the intersection of metabolic health in colocalized EA and W subjects. We include this in our discussion:
“Due to the investment of resources into ensuring a high level of phenotypic information on each cohort member, and due to its restricted geographical catchment area, the IDEO cohort was relatively small at the time of this analysis (n=46 individuals). This study only focused on two of the major ethnicities in the San Francisco Bay Area; as IDEO continues to expand and diversify its membership, we hope to study a sufficient number of participants from other ethnic groups in the future.”
The microbiome transfers from humans to mice also demonstrate that certain features of interpersonal or ethnic-related differences can be established in mice. This is useful for future studies, but it is not unexpected in and of itself given the robustness of transferring microbiome differences in other human-to-mouse studies. If the phenotype data were more compelling, then the utility of these transfers could be valuable.
We respectfully disagree with this point. To our knowledge, this is the first study demonstrating that ethnicity-associated differences in the gut microbiota are stable following transplantation, which is certainly not guaranteed given the marked and currently unpredictable variations between donor and recipient microbiotas shown here and in prior studies by us (Nayak et al., 2021; Turnbaugh et al., 2009b) and others (Walter et al., 2020).
We state this rationale in our results section:
“Taken together, our results support the hypothesis that there are stable ethnicity-associated signatures within the gut microbiota of lean EA vs. W individuals that are independent of diet. To experimentally test this hypothesis, we transplanted the gut microbiotas of two representative lean W and lean EA individuals into germ-free male C57BL/6J mice…Next, we sought to assess the reproducibility of these findings across multiple donors and in the context of a distinctive dietary pressure. We fed 20 germ-free male mice a high-fat, high-sugar (HFHS) diet for 4 weeks prior to colonization with a gut microbiota from one of 5 W and 5 EA donors....”
Furthermore, while the phenotypic data may not be as dramatic as the reviewer had hoped, this is to our knowledge the first demonstration that ethnicity-associated differences in the gut microbiota play a causal role in host phenotypes, as highlighted in our discussion:
“Our results in humans and mouse models support the broad potential for downstream consequences of ethnicity-associated differences in the gut microbiome for metabolic syndrome and potentially other disease areas. However, the causal relationships and how they can be understood in the context of the broader differences in host phenotype between ethnicities require further study.”
However, in the current state, I am concerned with the experimental design since the LFPP experiments used N=1 donor per ethnicity for establishing the mice colonies and are resultantly confounded by mice pseudo-replication with recipient mice derived from one donor of each ethnicity. This concern is relevant to interpreting results back to interpersonal or interethnic variation. Are phenotypic differences due to individual differences or ethnic differences? It's not clear.
We presented our data in summary form integrating the results from 3 independent experiments across two figures. To account for pseudoreplication as the reviewer suggests, we have restricted permutational space to account for one donor for multiple recipient mice using the parameters outlined in the adonis software package. Analyzing our results from 3 separate experiments, our results are statistically significant, which we mention in the revised text:
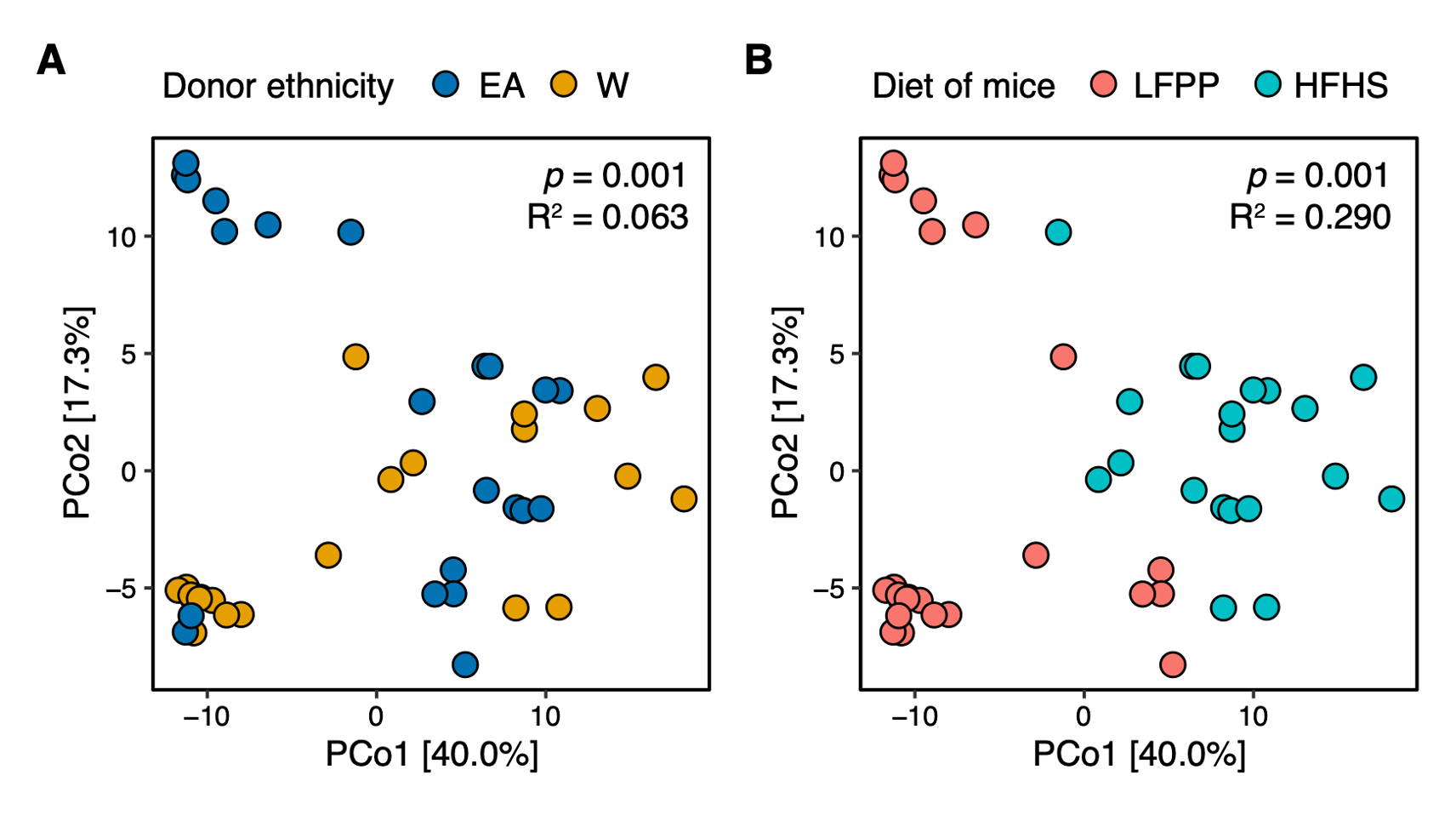
“In a pooled analysis of all gnotobiotic experiments accounting for one donor for multiple recipient mice, ethnicity and diet were both significantly associated with variations in the gut microbiota (Fig. S9), consistent with the extensive published data demonstrating the rapid and reproducible impact of a HFHS diet on the mouse and human gut microbiota (Bisanz et al., 2019).”
Figure S9. Combined analysis of recipient mice reveals significant associations with donor ethnicity and recipient diet. A PhILR PCoA is plotted based on 16S-Seq data from all gnotobiotic experiments. Individual mice are colored by (A) donor ethnicity or (B) the recipient’s diet. Both ethnicity and diet were statistically significant contributors to variance (ADONIS p-values and estimated variance displayed using blocks restricted by donor identifiers to account for one donor going to multiple recipient mice). We also observed a trend for interaction between diet and ethnicity in this model (p=0.068, R2=0.047, ADONIS).
The HFHS experiment also used N=5 donors that somewhat mitigates these concerns, but mixed sexes were used here and there can be sex-specific human microbiome differences.
Our study was designed to evaluate ethnicity and metabolic health. As we report in our original and updated analysis, we found no significant associations between the gut microbiota and biological sex (Figs. 2E and S4) in the IDEO cohort, perhaps due to the small effect size of sex reported in prior studies by other groups (Arumugam et al., 2011; Ding and Schloss, 2014; Schnorr et al., 2014; Zhang et al., 2021) coupled to the limited size of the current IDEO cohort.
The Turnbaugh and Koliwad labs use mixed sexes as donors for studies in conventionally raised and gnotobiotic mice due to our active funding from the NIH, which has clear guidelines meant to prevent continued discrimination against studies in females. The following link has additional information for your consideration: https://orwh.od.nih.gov/sex-gender/nih-policy-sex-biological-variable.
Importantly, our study was not confounded by sex due to the use of similar numbers of male and female donors (2 male and 2 females in the LFPP experiments and 3 female and 2 males for both ethnicities in the HFHS experiment). All of our recipient mice were male, as specified in our methods section and our revised main text:
“To experimentally test this hypothesis, we transplanted the gut microbiotas of two representative lean W and lean EA individuals into germ-free male C57BL/6J mice…Next, we sought to assess the reproducibility of these findings across multiple donors and in the context of a distinctive dietary pressure. We fed 20 germ-free male mice a high-fat, high-sugar (HFHS) diet for 4 weeks prior to colonization with a gut microbiota from one of 5 W and 5 EA donors....”
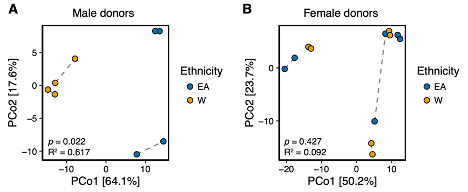
To further investigate any potential sex-specific signal we have stratified our analysis for the HFHS experiment by the gender of the donors (Reviewer Figure 2). This reveals that the significance between ethnicity in the microbiota transplantation experiments is preserved in mice that received stool from male donors (Reviewer Fig. 2A) but not female donors (Reviewer Fig. 2B). In Reviewer Fig. 1 above, LFPP1 and LFPP2 were conducted using different donors of different biological sex. Splitting our LFPP experiments up revealed the consistent signal for ethnicity in microbial community composition that we report above. The small sample sizes in this stratified analysis makes it difficult to conclude that there are reproducible sex-specific differences in the microbiome transplant experiments, but we agree with the reviewer that this question should be more thoroughly explored in future work.
We have added a brief note to the discussion to emphasize this important point:
“...differences between the human donor and recipient mouse microbiotas inherent to gnotobiotic transplantation warrant further investigation as do differences in the stability of the gut microbiotas of male versus female donors”
Reviewer Figure 2. (A,B) Principal coordinate analysis of PhILR Euclidean distances of stool from germ-free recipient mice transplanted with stool microbial communities from (A) male (n=2 EA and n=2 W donors) or (B) female (n=3 EA and n=3 W) donors of either ethnicity and fed a HFHS diet. Significance was assessed by ADONIS. Pairs of germ-free mice receiving the same donor sample are connected by a dashed line (n=2 recipient mice per donor). Experimental designs are shown in Fig. S7.
Finally, experimental results are not always consistent and sometimes show opposite trends that may be related to the sampling sizes. For instance, fat and lean mass increased and decreased respectively in LFPP, but there were no statistically-similar differences in HFHS. Moreover, the metabolic fat mass outcomes in mice do not match the expected human donor data. For instance, in LFPP1, White subjects had lower fat mass in humans but recipient mice on average gained more fat. It is difficult to reconcile these differences to a biological or sampling scheme reason.
We wholeheartedly agree with this point and were also surprised that the recipient mouse phenotypes did not match our original hypothesis based upon the observed health disparities between EA and W individuals. These surprising and perhaps counter-intuitive results demand further study and mechanistic dissection. We have tried to capture potential explanations for these findings while highlighting the limitations of our current study in our expanded discussion. With respect to the glucose tolerance data, the lack of a microbiome-driven phenotype might be due to the use of genetically identical mice that are not prone to metabolic illness without significant perturbation. If we had used mice prone to metabolic disease, such as non-obese diabetic (NOD) germ free recipient mice where the microbiome is known to impact the development of diabetes, we may have seen between ethnic differences in glucose tolerance.
Our revised discussion, with key points underlined is copied below for your convenience:
“Our results in humans and mouse models support the broad potential for downstream consequences of ethnicity-associated differences in the gut microbiome for metabolic syndrome and potentially other disease areas. However, the causal relationships and how they can be understood in the context of the broader differences in host phenotype between ethnicities require further study. While these data are consistent with our general hypothesis that ethnicity-associated differences in the gut microbiome are a source of differences in host metabolic disease risk, we were surprised by both the nature of the microbiome shifts and their directionality. Based upon observations in the IDEO (Alba et al., 2018) and other cohorts (Gu et al., 2006; Zheng et al., 2011), we anticipated that the gut microbiomes of lean EA individuals would promote obesity or other features of metabolic syndrome. In humans, we did find multiple signals that have been previously linked to obesity and its associated metabolic diseases in EA individuals, including increased Firmicutes (Basolo et al., 2020; Bisanz et al., 2019), decreased A. muciniphila (Depommier et al., 2019; Plovier et al., 2017), decreased diversity (Turnbaugh et al., 2009a), and increased acetate (Perry et al., 2016; Turnbaugh et al., 2006). Yet EA subjects also had higher levels of Bacteroidota and Bacteroides, which have been linked to improved metabolic health (Johnson et al., 2017). More importantly, our microbiome transplantations demonstrated that the recipients of the lean EA gut microbiome had less body fat despite consuming the same diet. These seemingly contradictory findings may suggest that the recipient mice lost some of the microbial features of ethnicity relevant to host metabolic disease or alternatively that the microbiome acts in a beneficial manner to counteract other ethnicity-associated factors driving disease.
EA subjects also had elevated levels of the short-chain fatty acids propionate and isobutyrate. The consequences of elevated intestinal propionate levels are unclear given the seemingly conflicting evidence in the literature that propionate may either exacerbate (Tirosh et al., 2019) or protect from (Lu et al., 2016) aspects of metabolic syndrome. Clinical data suggests that circulating propionate may be more relevant for disease than fecal levels (Müller et al., 2019), emphasizing the importance of considering both the specific microbial metabolites produced, their intestinal absorption, and their distribution throughout the body. Isobutyrate is even less well-characterized, with prior links to dietary intake (Berding and Donovan, 2018) but no association with obesity (Kim et al., 2019). Unlike SCFAs, we did not identify consistent differences in BCAAs, potentially due to differences in both extraction and standardization techniques inherent to GC-MS and NMR analysis (Cai et al., 2016; Lynch and Adams, 2014; Qin et al., 2012).
There are multiple limitations of this study. Due to the investment of resources into ensuring a high level of phenotypic information on each cohort member coupled to the restricted geographical catchment area, the IDEO cohort was relatively small at the time of this analysis (n=46 individuals). The current study only focused on two of the major ethnicities in the San Francisco Bay Area. As IDEO continues to expand and diversify its membership, we hope to study a sufficient number of participants from other ethnic groups. Stool samples were collected at a single time point and analyzed in a cross-sectional manner. While we used validated tools from the field of nutrition to monitor dietary intake, we cannot fully exclude subtle dietary differences between ethnicities (Johnson et al., 2019), which could be interrogated through controlled feeding studies (Basolo et al., 2020). Our mouse experiments were all performed in wild-type adult males. The use of a microbiome-dependent transgenic mouse model of diabetes (Brown et al., 2016) would be useful to test the effects of inter-ethnic differences in the microbiome on insulin and glucose tolerance. Additional experiments are warranted using the same donor inocula to colonize germ-free mice prior to concomitant feeding of multiple diets, allowing a more explicit test of the hypothesis that diet can disrupt ethnicity-associated microbial signatures. These studies, coupled to controlled experimentation with individual strains or more complex synthetic communities, would help to elucidate the mechanisms responsible for ethnicity-associated changes in host physiology and their relevance to disease.”
Reviewer #3 (Public Review):
The authors aimed to characterise how gut microbiota changes between different ethnic group for bacterial richness and community structure. They also wanted to address how this is associated with ethnic group within a defined geographical location. They have started to their story by comparing the fecal microbiota of relatively small cohort consisting of 46 lean and obese East Asian and White participants living in the San Francisco Bay Area. For that reason they used 16S and shotgun metagenomics. They demonstrated that ethnicity-associated differences in the gut microbiota are stronger in lean individuals and obese did not have a clear difference in the gut microbiota profile between ethnic groups, either suggesting that established obesity or its associated dietary patterns can overwrite long-lasting microbial signatures or alternatively that there is a shared ethnicity-independent microbiome type that predisposes individuals to obesity. The authors did also show the metabolic differences between these ethnic groups and the major differences were in the branched chain amino acid and the short-chain fatty acids. To prove their point, at this stage they have also used different metabolomic methodology. Although some aspects of the work are not very novel, the work does provide additional insights into the effect(s) of ethnicity, current living location and diet on shaping microbiota. Honestly, while reading through the manuscript, I have several questions where I believed that clarification was needed. But somehow, I felt like the authors have been reading my mind every step of the way. At the end of each section whatever I questioned was addressed in the next paragraph There are, however, a few points that I think would like to hear the authors' clarification.
- The authors pursued the story using 16S data. However, they have shotgun Metagenomics data which gives more power and resolution to microbiota profile. Is there any specific reason why the story was not build with shotgun Metagenomic data? However, if this is the case it will be nice to justify in the text or legend which figure was built with what dataset exactly?
As discussed above, 16S rRNA gene and metagenomic sequencing both have strengths and weaknesses. For example, 16S-seq is inexpensive and allows analysis of low abundance species, whereas metagenomics permits analysis of gene and pathway abundances of abundant taxa. As requested, we have now expanded Figure 2 (metagenomics) to better match Figure 1 (16S-seq). The type of technology is defined within each legend and the relevant text within our results.
- Even though the authors mentioned in the discussion that they have not used the same inocula from a donor to different diet, it will be nice if the authors further comments whether they would expect the same results or slightly different results which each different inocula.
As requested, we have modified the text in our discussion to include these comments:
“Additional experiments are warranted using the same donor inocula to colonize germ-free mice prior to concomitant feeding of multiple diets, allowing a more explicit test of the hypothesis that diet can disrupt ethnicity-associated microbial signatures. These studies, coupled to controlled experimentation with individual strains or more complex synthetic communities, would help to elucidate the mechanisms responsible for ethnicity-associated changes in host physiology and their relevance to disease.”
Overall, the study is well executed and claims and conclusions seem relatively well justified by the provided evidence. The findings are interesting for a broad audience of biologists. The findings are interesting for a broad audience of biologists.
-

Evaluation Summary:
This study aims to measure interpersonal and interethnic variation in human microbiomes and metabolomes in the San Franciso, CA, area. The strength of the study is in the level of robust analysis of the microbiota. It is interesting that diet is not one of the apparent associations in this study, yet the relationship of microbiota diversity to body habitus is strong in Caucasian subjects. Overall, the key results of this work nicely confirm that there are dissimilarities in gut microbiomes related to differences in ethnicity of subjects.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #3 agreed to share their name with the authors.)
-

Reviewer #1 (Public Review):
The key question that the authors were addressing was how ethnicity differentially affects the microbiota of subjects living in a particular area (in this case East Asians and Caucasians living in San Francisco that have been enrolled in an 'Inflammation, Diabetes, Ethnicity and Obesity cohort - although inflammatory disease was apparently excluded in these subjects).
The existence of differences between different populations allows potential discrimination of the underlying factors - such as host genetics, diet, lifestyle, physiological parameters, body habitus or other environmental influences. In this case body habitus has been selected as a stratification factor between the two ethnicities. Immigration potentially allows distinction of environmental and host genetical influences.
The strength of the …Reviewer #1 (Public Review):
The key question that the authors were addressing was how ethnicity differentially affects the microbiota of subjects living in a particular area (in this case East Asians and Caucasians living in San Francisco that have been enrolled in an 'Inflammation, Diabetes, Ethnicity and Obesity cohort - although inflammatory disease was apparently excluded in these subjects).
The existence of differences between different populations allows potential discrimination of the underlying factors - such as host genetics, diet, lifestyle, physiological parameters, body habitus or other environmental influences. In this case body habitus has been selected as a stratification factor between the two ethnicities. Immigration potentially allows distinction of environmental and host genetical influences.
The strength of the study is in the level of robust analysis of the microbiotas by a very experienced group of researchers, distinguishing the microbiota differences, especially in lean subject, with analysis of associations that may be driving the differences. It is interesting that diet is not one of the apparent associations in this study, yet the relationship of microbiota diversity to body habitus is strong in Caucasian subjects. These associations cannot easily be extrapolated to causation or mechanism - a fact well recognized in the paper - but remain important observations that rationalize in vivo modeling with experimental animals or in vitro analyses of microbial interactions between different taxa simulating the context of differences in the intestinal milieu. The paper includes work showing that differences of the microbiota can be recapitulated after transfer to germ-free mice, at least over the short term: this is important to provide tools to model the reasons for differences in consortial composition.
A very large amount of work required to assemble the samples and the clinical phenotypic metadata set making the data an important and definitive contribution for the subjects studied. Of course, this is one sample of extremely variable human conditions and lifestyles that will help build the overall picture of how differences in our genetics and environment shape our intestinal microbiota. -

Reviewer #2 (Public Review):
The study's primary aims are to test for the differences in the microbiome between self-identified East Asian and White subjects from the San Francisco area in the new IDEO cohort. The study builds on an growing literature which describes variations among ethnic groups. The major conclusion of "emphasize the utility of studying diverse ethnic groups" is not novel to the literature.
Overall, the strength of the results is that they confirm patterns from different cohorts/studies and demonstrate that ethnic-related differences are common. The results are subject to sample size concerns that may underpin some of the conflicting or lack of significant results. For instance, there is no overlap in highlighted species-level taxonomy differences between 16S and metagenomic analyses, which precludes a clear …
Reviewer #2 (Public Review):
The study's primary aims are to test for the differences in the microbiome between self-identified East Asian and White subjects from the San Francisco area in the new IDEO cohort. The study builds on an growing literature which describes variations among ethnic groups. The major conclusion of "emphasize the utility of studying diverse ethnic groups" is not novel to the literature.
Overall, the strength of the results is that they confirm patterns from different cohorts/studies and demonstrate that ethnic-related differences are common. The results are subject to sample size concerns that may underpin some of the conflicting or lack of significant results. For instance, there is no overlap in highlighted species-level taxonomy differences between 16S and metagenomic analyses, which precludes a clear interpretation of the meaning of those differences and whether taxa should be highlighted in the abstract; there are low AUC values for the random forest modelling; and there is a lack of significance in correlations between BMI and East Asian subjects in F4a where there may be a correlation. While a minor point, it serves to highlight the sample sizes as the range of the variation in East Asian subjects is not as substantial as the White subjects because there are fewer East Asian data points above a 30 BMI (~N=5) relative to those of White subjects (~N=11).
The microbiome transfers from humans to mice also demonstrate that certain features of interpersonal or ethnic-related differences can be established in mice. This is useful for future studies, but it is not unexpected in and of itself given the robustness of transferring microbiome differences in other human-to-mouse studies. If the phenotype data were more compelling, then the utility of these transfers could be valuable. However, in the current state, I am concerned with the experimental design since the LFPP experiments used N=1 donor per ethnicity for establishing the mice colonies and are resultantly confounded by mice pseudo-replication with recipient mice derived from one donor of each ethnicity. This concern is relevant to interpreting results back to interpersonal or interethnic variation. Are phenotypic differences due to individual differences or ethnic differences? It's not clear. The HFHS experiment also used N=5 donors that somewhat mitigates these concerns, but mixed sexes were used here and there can be sex-specific human microbiome differences. Finally, experimental results are not always consistent and sometimes show opposite trends that may be related to the sampling sizes. For instance, fat and lean mass increased and decreased respectively in LFPP, but there were no statistically-similar differences in HFHS. Moreover, the metabolic fat mass outcomes in mice do not match the expected human donor data. For instance, in LFPP1, White subjects had lower fat mass in humans but recipient mice on average gained more fat. It is difficult to reconcile these differences to a biological or sampling scheme reason.
-

Reviewer #3 (Public Review):
The authors aimed to characterise how gut microbiota changes between different ethnic group for bacterial richness and community structure. They also wanted to address how this is associated with ethnic group within a defined geographical location. They have started to their story by comparing the fecal microbiota of relatively small cohort consisting of 46 lean and obese East Asian and White
participants living in the San Francisco Bay Area. For that reason they used 16S and shotgun metagenomics. They demonstrated that ethnicity-associated differences in the gut microbiota are stronger in lean individuals and obese did not have a clear difference in the gut microbiota profile between ethnic groups, either suggesting that established obesity or its associated dietary patterns can overwrite long-lasting …Reviewer #3 (Public Review):
The authors aimed to characterise how gut microbiota changes between different ethnic group for bacterial richness and community structure. They also wanted to address how this is associated with ethnic group within a defined geographical location. They have started to their story by comparing the fecal microbiota of relatively small cohort consisting of 46 lean and obese East Asian and White
participants living in the San Francisco Bay Area. For that reason they used 16S and shotgun metagenomics. They demonstrated that ethnicity-associated differences in the gut microbiota are stronger in lean individuals and obese did not have a clear difference in the gut microbiota profile between ethnic groups, either suggesting that established obesity or its associated dietary patterns can overwrite long-lasting microbial signatures or alternatively that there is a shared ethnicity-independent microbiome type that predisposes individuals to obesity. The authors did also show the metabolic differences between these ethnic groups and the major differences were in the branched chain amino acid and the short-chain fatty acids. To prove their point, at this stage they have also used different metabolomic methodology. Although some aspects of the work are not very novel, the work does provide additional insights into the effect(s) of ethnicity, current living location and diet on shaping microbiota. Honestly, while reading through the manuscript, I have several questions where I believed that clarification was needed. But somehow, I felt like the authors have been reading my mind every step of the way. At the end of each section whatever I questioned was addressed in the next paragraph There are, however, a few points that I think would like to hear the authors' clarification.- The authors pursued the story using 16S data. However, they have shotgun Metagenomics data which gives more power and resolution to microbiota profile. Is there any specific reason why the story was not build with shotgun Metagenomic data? However, if this is the case it will be nice to justify in the text or legend which figure was built with what dataset exactly?
- Even though the authors mentioned in the discussion that they have not used the same inocula from a donor to different diet, it will be nice if the authors further comments whether they would expect the same results or slightly different results which each different inocula.Overall, the study is well executed and claims and conclusions seem relatively well justified by the provided evidence. The findings are interesting for a broad audience of biologists. The findings are interesting for a broad audience of biologists.
-