Prediction of type 2 diabetes mellitus onset using logistic regression-based scorecards
Curation statements for this article:-
Curated by eLife
Evaluation Summary:
The authors have used the UK bio-bank with sophisticated statistical modeling to predict the risk of type 2 diabetes mellitus development. Prognosis and early detection of diabetes are key factors in clinical practice and the current data suggest a new machine-learning based algorithm that further advances our ability to prevent diabetes.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #1 and Reviewer #3 agreed to share their name with the authors.)
This article has been Reviewed by the following groups
Discuss this preprint
Start a discussion What are Sciety discussions?Listed in
- Evaluated articles (eLife)
Abstract
Type 2 diabetes (T2D) accounts for ~90% of all cases of diabetes, resulting in an estimated 6.7 million deaths in 2021, according to the International Diabetes Federation. Early detection of patients with high risk of developing T2D can reduce the incidence of the disease through a change in lifestyle, diet, or medication. Since populations of lower socio-demographic status are more susceptible to T2D and might have limited resources or access to sophisticated computational resources, there is a need for accurate yet accessible prediction models.
Methods:
In this study, we analyzed data from 44,709 nondiabetic UK Biobank participants aged 40–69, predicting the risk of T2D onset within a selected time frame (mean of 7.3 years with an SD of 2.3 years). We started with 798 features that we identified as potential predictors for T2D onset. We first analyzed the data using gradient boosting decision trees, survival analysis, and logistic regression methods. We devised one nonlaboratory model accessible to the general population and one more precise yet simple model that utilizes laboratory tests. We simplified both models to an accessible scorecard form, tested the models on normoglycemic and prediabetes subcohorts, and compared the results to the results of the general cohort. We established the nonlaboratory model using the following covariates: sex, age, weight, height, waist size, hip circumference, waist-to-hip ratio, and body mass index. For the laboratory model, we used age and sex together with four common blood tests: high-density lipoprotein (HDL), gamma-glutamyl transferase, glycated hemoglobin, and triglycerides. As an external validation dataset, we used the electronic medical record database of Clalit Health Services.
Results:
The nonlaboratory scorecard model achieved an area under the receiver operating curve (auROC) of 0.81 (95% confidence interval [CI] 0.77–0.84) and an odds ratio (OR) between the upper and fifth prevalence deciles of 17.2 (95% CI 5–66). Using this model, we classified three risk groups, a group with 1% (0.8–1%), 5% (3–6%), and the third group with a 9% (7–12%) risk of developing T2D. We further analyzed the contribution of the laboratory-based model and devised a blood test model based on age, sex, and the four common blood tests noted above. In this scorecard model, we included age, sex, glycated hemoglobin (HbA1c%), gamma glutamyl-transferase, triglycerides, and HDL cholesterol. Using this model, we achieved an auROC of 0.87 (95% CI 0.85–0.90) and a deciles' OR of ×48 (95% CI 12–109). Using this model, we classified the cohort into four risk groups with the following risks: 0.5% (0.4–7%); 3% (2–4%); 10% (8–12%); and a high-risk group of 23% (10–37%) of developing T2D. When applying the blood tests model using the external validation cohort (Clalit), we achieved an auROC of 0.75 (95% CI 0.74–0.75). We analyzed several additional comprehensive models, which included genotyping data and other environmental factors. We found that these models did not provide cost-efficient benefits over the four blood test model. The commonly used German Diabetes Risk Score (GDRS) and Finnish Diabetes Risk Score (FINDRISC) models, trained using our data, achieved an auROC of 0.73 (0.69–0.76) and 0.66 (0.62–0.70), respectively, inferior to the results achieved by the four blood test model and by the anthropometry models.
Conclusions:
The four blood test and anthropometric models outperformed the commonly used nonlaboratory models, the FINDRISC and the GDRS. We suggest that our models be used as tools for decision-makers to assess populations at elevated T2D risk and thus improve medical strategies. These models might also provide a personal catalyst for changing lifestyle, diet, or medication modifications to lower the risk of T2D onset.
Funding:
The funders had no role in study design, data collection, interpretation, or the decision to submit the work for publication.
Article activity feed
-

-

Author Response
Reviewer #3 (Public Review):
The authors analyzed several models for predicting the early onset of T2D, where they trained and tested on a UKB based cohort, aged 40 - 69 and suggest two simple logistic regression models: the anthropometric and the five blood tests models in reference to FINDRISC and GDRS models. Their models achieved better auROC, APS, and decile prevalence OR, and better-calibrated predictions.
Strengths:
1.The authors have neatly explained their objectives and performed well-justified analyses.
2.The authors highlight how using both features - HbA1C% measure and reticulocyte count may provide a better indication of the average blood sugar level during the last two-three months than using just the standard HbA1C% measure.
3.Further verification of the proposed anthropometric-based and 5 …
Author Response
Reviewer #3 (Public Review):
The authors analyzed several models for predicting the early onset of T2D, where they trained and tested on a UKB based cohort, aged 40 - 69 and suggest two simple logistic regression models: the anthropometric and the five blood tests models in reference to FINDRISC and GDRS models. Their models achieved better auROC, APS, and decile prevalence OR, and better-calibrated predictions.
Strengths:
1.The authors have neatly explained their objectives and performed well-justified analyses.
2.The authors highlight how using both features - HbA1C% measure and reticulocyte count may provide a better indication of the average blood sugar level during the last two-three months than using just the standard HbA1C% measure.
3.Further verification of the proposed anthropometric-based and 5 blood-test results-based modelscan discriminate discriminating within a group of normoglycemic participants and within a group of pre-diabetic participants resulted in outperforming the FINDRISC and the GDRS based models.
Weaknesses:
- As the authors point out in the manuscript that these models are suited for the UKB cohort or populations with similar characteristics. It limits the extrapolation of these findings onto another cohort from a different background until analyzed on another country/continent-based cohort.
We agree with this comment as we indeed pointed in the paper. We recommend to adjust these models when applying it to populations with distinct characteristics.
- In the methods section, an additional explanation of how the T2D prevalence bins were formed would be useful to a reader.
We thank the reviewer for this note, we added the following explanation in section 4.11: “We considered several potential risk score limits that separate T2D onset probability in each of the scores groups, and we chose boundaries that showed a separation between the risk groups on the validation datasets. Once we decided on the boundaries of the score, we report the prevalence in each risk group on the test set and we report these results.”
- The authors have mentioned that the prevalence of diabetes has been rising more rapidly in low and middle-income countries (LMICs) than in high-income countries and the objective of the present research was to develop clinically usable models which are easy to use and highly predictive of T2D onset. As lifestyle is also one of the contributory factors for T2D, additional analysis that includes a comparison of groups between low-income and high-income subjects within UKB-based cohort provided such metadata available would help understand if the prevalence for T2D differs or not between such groups.
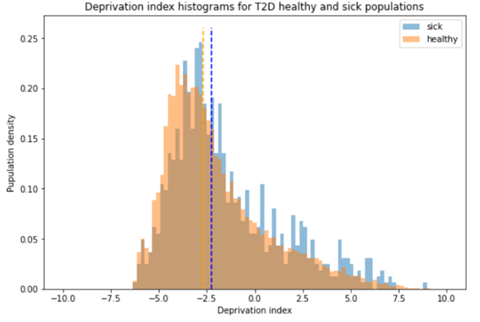
We thank the reviewer for this comment, we added below an analysis that we run on our data, showing the deprivation indexes differences between sick and healthy populations. The sick population has a higher deprivation index as expected. When running a Mann-Whitney U Test on the data we get a p value of zero, creating this with a sample of just 1000 participants from each group, we get a p-value of 2.37e-137. This indicates that there is a significant correlation between deprivation index and tendency to develop T2D. We also add this finding to the supplementary material and a reference to it.
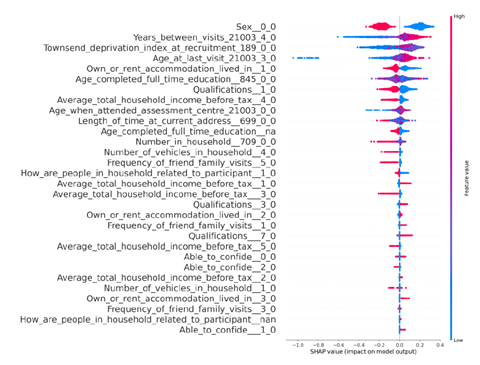
You can also find below a SHAP diagram showing tht higher Townsend deprivation index is pushing the prediction for T2D upwards.
-

Evaluation Summary:
The authors have used the UK bio-bank with sophisticated statistical modeling to predict the risk of type 2 diabetes mellitus development. Prognosis and early detection of diabetes are key factors in clinical practice and the current data suggest a new machine-learning based algorithm that further advances our ability to prevent diabetes.
(This preprint has been reviewed by eLife. We include the public reviews from the reviewers here; the authors also receive private feedback with suggested changes to the manuscript. Reviewer #1 and Reviewer #3 agreed to share their name with the authors.)
-

Reviewer #1 (Public Review):
The authors succeeded in providing data-based models (including both lab and non-lab data) to predict the risk of diabetes type 2 development. The major strength of this manuscript is the sound methodology and robust statistical methods used. However, the link between the developed items with more clinical aspects of such tools is partly missing in this manuscript and a further evaluation by and recruitment of the clinicians and epidemiologists would help enrich the proposed material. In this effort, authors almost reached their study aim; however, final improvements and amendments are needed to advantage the high-risk part of the population.
-

Reviewer #2 (Public Review):
Developing simple methods that can predict onset of T2D is an important research area. I think that transforming a machine learning model to a scorecard that can be used without computational resources has the potential to be very useful. However, my fear is that the authors have jumped the gun to early on this. Before the prediction model is clinically validated in a relevant cohort, it has little value in my opinion. This is a process that can be performed to a machine-learning model that has been validated in multiple settings, and now a scorecard can be implemented to ease it use.
The predictive results presented in this study are very strong - a highly simplistic model that is based only on age, sex and body measurements achieves AUC=0.82, a blood tests-based model with only 5 blood tests achieves 0.89. …
Reviewer #2 (Public Review):
Developing simple methods that can predict onset of T2D is an important research area. I think that transforming a machine learning model to a scorecard that can be used without computational resources has the potential to be very useful. However, my fear is that the authors have jumped the gun to early on this. Before the prediction model is clinically validated in a relevant cohort, it has little value in my opinion. This is a process that can be performed to a machine-learning model that has been validated in multiple settings, and now a scorecard can be implemented to ease it use.
The predictive results presented in this study are very strong - a highly simplistic model that is based only on age, sex and body measurements achieves AUC=0.82, a blood tests-based model with only 5 blood tests achieves 0.89. As far as I am aware, those are better than previous studies. However, since I didn't observe methodological advancements in the is study, these results raise doubts whether the analysis is consistent with previous studies, and we can really compare apples to apples.
-

Reviewer #3 (Public Review):
The authors analyzed several models for predicting the early onset of T2D, where they trained and tested on a UKB based cohort, aged 40 - 69 and suggest two simple logistic regression models: the anthropometric and the five blood tests models in reference to FINDRISC and GDRS models. Their models achieved better auROC, APS, and decile prevalence OR, and better-calibrated predictions.
Strengths:
1. The authors have neatly explained their objectives and performed well-justified analyses.
2. The authors highlight how using both features - HbA1C% measure and reticulocyte count may provide a better indication of the average blood sugar level during the last two-three months than using just the standard HbA1C% measure.
3. Further verification of the proposed anthropometric-based and 5 blood-test results-based …
Reviewer #3 (Public Review):
The authors analyzed several models for predicting the early onset of T2D, where they trained and tested on a UKB based cohort, aged 40 - 69 and suggest two simple logistic regression models: the anthropometric and the five blood tests models in reference to FINDRISC and GDRS models. Their models achieved better auROC, APS, and decile prevalence OR, and better-calibrated predictions.
Strengths:
1. The authors have neatly explained their objectives and performed well-justified analyses.
2. The authors highlight how using both features - HbA1C% measure and reticulocyte count may provide a better indication of the average blood sugar level during the last two-three months than using just the standard HbA1C% measure.
3. Further verification of the proposed anthropometric-based and 5 blood-test results-based models, i.e. if they are capable of discriminating within a group of normoglycemic participants and within a group of pre-diabetic participants resulted in outperforming the FINDRISC and the GDRS based models.
Weaknesses:
1. As the authors point out in the manuscript that these models are suited for the UKB cohort or populations with similar characteristics. It limits the extrapolation of these findings onto another cohort from a different background until analyzed on another country/continent-based cohort.
2. In the methods section, an additional explanation of how the T2D prevalence bins were formed would be useful to a reader.
3. The authors have mentioned that the prevalence of diabetes has been rising more rapidly in low and middle-income countries (LMICs) than in high-income countries and the objective of the present research was to develop clinically usable models which are easy to use and highly predictive of T2D onset. As lifestyle is also one of the contributory factors for T2D, additional analysis that includes a comparison of groups between low-income and high-income subjects within UKB-based cohort provided such metadata available would help understand if the prevalence for T2D differs or not between such groups.
Overall, the authors achieve their aims and the results clearly support their conclusions.
The data analyses performed on a large UKB cohort show promising outcomes, however, the research in its current form is on a pilot scale, if the suggested models work similarly better than the reference highly-esteemed control models on other country-based cohorts, the impact of this study will serve to be a useful tool for the clinicians and to a broader community.
-